APOE3 Christchurch modulates β-catenin/Wnt signaling in iPS cell-derived cerebral organoids from Alzheimer's cases
- PMID: 38571814
- PMCID: PMC10987717
- DOI: 10.3389/fnmol.2024.1373568
APOE3 Christchurch modulates β-catenin/Wnt signaling in iPS cell-derived cerebral organoids from Alzheimer's cases
Abstract
A patient with the PSEN1 E280A mutation and homozygous for APOE3 Christchurch (APOE3Ch) displayed extreme resistance to Alzheimer's disease (AD) cognitive decline and tauopathy, despite having a high amyloid burden. To further investigate the differences in biological processes attributed to APOE3Ch, we generated induced pluripotent stem (iPS) cell-derived cerebral organoids from this resistant case and a non-protected control, using CRISPR/Cas9 gene editing to modulate APOE3Ch expression. In the APOE3Ch cerebral organoids, we observed a protective pattern from early tau phosphorylation. ScRNA sequencing revealed regulation of Cadherin and Wnt signaling pathways by APOE3Ch, with immunostaining indicating elevated β-catenin protein levels. Further in vitro reporter assays unexpectedly demonstrated that ApoE3Ch functions as a Wnt3a signaling enhancer. This work uncovered a neomorphic molecular mechanism of protection of ApoE3 Christchurch, which may serve as the foundation for the future development of protected case-inspired therapeutics targeting AD and tauopathies.
Keywords: Alzheimer’s disease; ApoE; ApoE Christchurch; CRISPR; Presenilin; Wnt signaling; iPS cells.
Copyright © 2024 Perez-Corredor, Vanderleest, Vacano, Sanchez, Villalba-Moreno, Marino, Krasemann, Mendivil-Perez, Aguillón, Jiménez-Del-Río, Baena, Sepulveda-Falla, Lopera, Quiroz, Arboleda-Velasquez and Mazzarino.
Conflict of interest statement
JFA-V, YTQ, and FL are listed as inventors on a patent application addressing Christchurch-inspired therapeutics filed by Mass General Brigham. JFA-V is a co-founder of Epoch Biotech, a company developing ApoE Christchurch-inspired therapeutics. YTQ serves as a consultant for Biogen. FL received consulting fees from Biogen and Tecnoquimicas. GV is employed by the company Vacano Informatics LLC of Arvada, CO, USA and was contracted by JFA-V. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

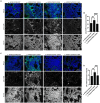

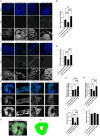


Update of
-
APOE3 Christchurch modulates tau phosphorylation and β-catenin/Wnt/Cadherin signaling in induced pluripotent stem cell-derived cerebral organoids from Alzheimer's cases.bioRxiv [Preprint]. 2023 Jan 13:2023.01.11.523290. doi: 10.1101/2023.01.11.523290. bioRxiv. 2023. Update in: Front Mol Neurosci. 2024 Mar 20;17:1373568. doi: 10.3389/fnmol.2024.1373568. PMID: 36712026 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

