Genomic insights into the population history and adaptive traits of Latin American Criollo cattle
- PMID: 38571912
- PMCID: PMC10990470
- DOI: 10.1098/rsos.231388
Genomic insights into the population history and adaptive traits of Latin American Criollo cattle
Abstract
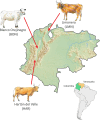
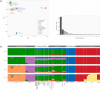
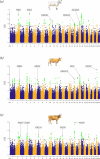
Criollo cattle, the descendants of animals brought by Iberian colonists to the Americas, have been the subject of natural and human-mediated selection in novel tropical agroecological zones for centuries. Consequently, these breeds have evolved distinct characteristics such as resistance to diseases and exceptional heat tolerance. In addition to European taurine (Bos taurus) ancestry, it has been proposed that gene flow from African taurine and Asian indicine (Bos indicus) cattle has shaped the ancestry of Criollo cattle. In this study, we analysed Criollo breeds from Colombia and Venezuela using whole-genome sequencing (WGS) and single-nucleotide polymorphism (SNP) array data to examine population structure and admixture at high resolution. Analysis of genetic structure and ancestry components provided evidence for African taurine and Asian indicine admixture in Criollo cattle. In addition, using WGS data, we detected selection signatures associated with a myriad of adaptive traits, revealing genes linked to thermotolerance, reproduction, fertility, immunity and distinct coat and skin coloration traits. This study underscores the remarkable adaptability of Criollo cattle and highlights the genetic richness and potential of these breeds in the face of climate change, habitat flux and disease challenges. Further research is warranted to leverage these findings for more effective and sustainable cattle breeding programmes.
Keywords: Latin America; cattle; microevolution; population genomics; selection; thermotolerance.
© 2024 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures

Similar articles
-
Whole genome characterization of autochthonous Bos taurus brachyceros and introduced Bos indicus indicus cattle breeds in Cameroon regarding their adaptive phenotypic traits and pathogen resistance.BMC Genet. 2020 Jun 22;21(1):64. doi: 10.1186/s12863-020-00869-9. BMC Genet. 2020. PMID: 32571206 Free PMC article.
-
Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle.PLoS Genet. 2014 Mar 27;10(3):e1004254. doi: 10.1371/journal.pgen.1004254. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24675901 Free PMC article.
-
The patterns of admixture, divergence, and ancestry of African cattle populations determined from genome-wide SNP data.BMC Genomics. 2020 Dec 7;21(1):869. doi: 10.1186/s12864-020-07270-x. BMC Genomics. 2020. PMID: 33287702 Free PMC article.
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Pre-pubertal and postpartum anestrus in tropical Zebu cattle.Anim Reprod Sci. 2004 Jul;82-83:373-87. doi: 10.1016/j.anireprosci.2004.05.006. Anim Reprod Sci. 2004. PMID: 15271467 Review.
References
-
- Rouse JE. 1977. The Criollo: Spanish cattle in the Americas. Norman, OK: University of Oklahoma Press.
-
- Armenteros Martínez I. 2018. The Canary Islands as an area of interconnectivity between the Mediterranean and the Atlantic (fourteenth-sixteenth centuries). In Entre mers—Outre-mer: spaces, modes and agents of Indo-Mediterranean connectivity (eds Jaspert N, Kolditz S), pp. 201–217, Heidelberg, Germany: Heidelberg University Publishing.
-
- Rodero A. 1992. Primitive Andalusian livestock and their implications in the discovery of America. Arch. Zootec. 41 , 383–400.
-
- Dunmire WW. 2004. Gardens of new Spain: how Mediterranean plants and foods changed America, 1st edn. Austin, TX: University of Texas Press.
-
- Crosby AW. 2003. The Columbian exchange: biological and cultural consequences of 1492, 30th anniversary edn. Westport, CT: Praeger.
Associated data
LinkOut - more resources
Full Text Sources

