Predicting immunotherapy response in melanoma using a novel tumor immunological phenotype-related gene index
- PMID: 38571962
- PMCID: PMC10987686
- DOI: 10.3389/fimmu.2024.1343425
Predicting immunotherapy response in melanoma using a novel tumor immunological phenotype-related gene index
Abstract
Introduction: Melanoma is a highly aggressive and recurrent form of skin cancer, posing challenges in prognosis and therapy prediction.
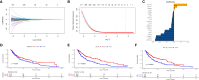
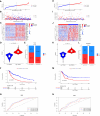
Methods: In this study, we developed a novel TIPRGPI consisting of 20 genes using Univariate Cox regression and the LASSO algorithm. The high and low-risk groups based on TIPRGPI exhibited distinct mutation profiles, hallmark pathways, and immune cell infiltration in the tumor microenvironment.
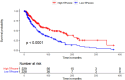
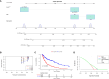
Results: Notably, significant differences in tumor immunogenicity and TIDE were observed between the risk groups, suggesting a better response to immune checkpoint blockade therapy in the low-TIPRGPI group. Additionally, molecular docking predicted 10 potential drugs that bind to the core target, PTPRC, of the TIPRGPI signature.
Discussion: Our findings highlight the reliability of TIPRGPI as a prognostic signature and its potential application in risk classification, immunotherapy response prediction, and drug candidate identification for melanoma treatment. The "TIP genes" guided strategy presented in this study may have implications beyond melanoma and could be applied to other cancer types.
Keywords: bioinformatics; immunotherapy response; melanoma; molecular docking; prognosis; tumor microenvironment.
Copyright © 2024 Zheng, He, Chen, Gu, Wei, Chen and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Identification of a Tumor Immunological Phenotype-Related Gene Signature for Predicting Prognosis, Immunotherapy Efficacy, and Drug Candidates in Hepatocellular Carcinoma.Front Immunol. 2022 Apr 12;13:862527. doi: 10.3389/fimmu.2022.862527. eCollection 2022. Front Immunol. 2022. PMID: 35493471 Free PMC article.
-
The development and experimental validation of hypoxia-related long noncoding RNAs prognostic signature in predicting prognosis and immunotherapy of cutaneous melanoma.Aging (Albany NY). 2023 Nov 2;15(21):11918-11939. doi: 10.18632/aging.205157. Epub 2023 Nov 2. Aging (Albany NY). 2023. PMID: 37921852 Free PMC article.
-
Development and validation of an immune gene set-based prognostic signature in cutaneous melanoma.Future Oncol. 2021 Nov;17(31):4115-4129. doi: 10.2217/fon-2021-0104. Epub 2021 Jul 22. Future Oncol. 2021. PMID: 34291650
-
Identification of immune activation-related gene signature for predicting prognosis and immunotherapy efficacy in lung adenocarcinoma.Front Immunol. 2023 Jul 7;14:1217590. doi: 10.3389/fimmu.2023.1217590. eCollection 2023. Front Immunol. 2023. PMID: 37492563 Free PMC article.
-
DNA damage repair-related gene signature predicts prognosis and indicates immune cell infiltration landscape in skin cutaneous melanoma.Front Endocrinol (Lausanne). 2022 Jul 26;13:882431. doi: 10.3389/fendo.2022.882431. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35957812 Free PMC article.
Cited by
-
Unraveling the role of histone acetylation in sepsis biomarker discovery.Front Mol Biosci. 2025 Apr 30;12:1582181. doi: 10.3389/fmolb.2025.1582181. eCollection 2025. Front Mol Biosci. 2025. PMID: 40370519 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

