Blockade of the deubiquitinating enzyme USP48 degrades oncogenic HMGA2 and inhibits colorectal cancer invasion and metastasis
- PMID: 38572092
- PMCID: PMC10985028
- DOI: 10.1016/j.apsb.2024.01.006
Blockade of the deubiquitinating enzyme USP48 degrades oncogenic HMGA2 and inhibits colorectal cancer invasion and metastasis
Abstract
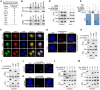
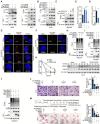
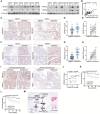
HMGA2, a pivotal transcription factor, functions as a versatile regulator implicated in the progression of diverse aggressive malignancies. In this study, mass spectrometry was employed to identify ubiquitin-specific proteases that potentially interact with HMGA2, and USP48 was identified as a deubiquitinating enzyme of HMGA2. The enforced expression of USP48 significantly increased HMGA2 protein levels by inhibiting its degradation, while the deprivation of USP48 promoted HMGA2 degradation, thereby suppressing tumor invasion and metastasis. We discovered that USP48 undergoes SUMOylation at lysine 258, which enhances its binding affinity to HMGA2. Through subsequent phenotypic screening of small molecules, we identified DUB-IN-2 as a remarkably potent pharmacological inhibitor of USP48. Interestingly, the small-molecule inhibitor targeting USP48 induces destabilization of HMGA2. Clinically, upregulation of USP48 or HMGA2 in cancerous tissues is indicative of poor prognosis for patients with colorectal cancer (CRC). Collectively, our study not only elucidates the regulatory mechanism of DUBs involved in HMGA2 stability and validates USP48 as a potential therapeutic target for CRC, but also identifies DUB-IN-2 as a potent inhibitor of USP48 and a promising candidate for CRC treatment.
Keywords: Colorectal cancer; HMGA2; Invasion and metastasis; Post-translational modification; SUMOylation; Specific inhibitors; USP48; Ubiquitination.
© 2024 The Authors.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures






References
-
- Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. - PubMed
-
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136:E359–E386. - PubMed
-
- Hoeller D., Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. - PubMed
LinkOut - more resources
Full Text Sources

