Comparative analysis of the gut bacteria and fungi in migratory demoiselle cranes (Grus virgo) and common cranes (Grus grus) in the Yellow River Wetland, China
- PMID: 38572234
- PMCID: PMC10987826
- DOI: 10.3389/fmicb.2024.1341512
Comparative analysis of the gut bacteria and fungi in migratory demoiselle cranes (Grus virgo) and common cranes (Grus grus) in the Yellow River Wetland, China
Abstract
Introduction: Gut microbiota are closely related to the nutrition, immunity, and metabolism of the host and play important roles in maintaining the normal physiological activities of animals. Cranes are important protected avian species in China, and they are sensitive to changes in the ecological environment and are thus good environmental indicators. There have been no reports examining gut fungi or the correlation between bacteria and fungi in wild Demoiselle cranes (Grus virgo) and Common cranes (Grus grus). Related research can provide a foundation for the protection of rare wild animals.
Methods: 16S rRNA and ITS high-throughput sequencing techniques were used to analyze the gut bacterial and fungal diversity of Common and Demoiselle cranes migrating to the Yellow River wetland in Inner Mongolia.
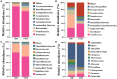
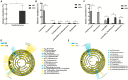
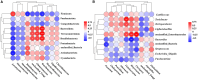
Results: The results revealed that for gut bacteria α diversity, Chao1 index in Demoiselle cranes was remarkably higher than that in Common cranes (411.07 ± 79.54 vs. 294.92 ± 22.38), while other index had no remarkably differences. There was no remarkable difference in fungal diversity. There were marked differences in the gut microbial composition between the two crane species. At the phylum level, the highest abundance of bacteria in the Common crane and Demoiselle crane samples was Firmicutes, accounting for 87.84% and 74.29%, respectively. The highest abundance of fungi in the guts of the Common and Demoiselle cranes was Ascomycota, accounting for 69.42% and 57.63%, respectively. At the genus level, the most abundant bacterial genus in the Common crane sample was Turicibacter (38.60%), and the most abundant bacterial genus in the Demoiselle crane sample was Catelicoccus (39.18%). The most abundant fungi in the Common crane sample was Penicillium (6.97%), and the most abundant fungi in the Demoiselle crane sample was Saccharomyces (8.59%). Correlation analysis indicated that there was a significant correlation between gut bacteria and fungi.
Discussion: This study provided a research basis for the protection of cranes. Indeed, a better understanding of the gut microbiota is very important for the conservation and management of wild birds, as it not only helps us to understand their life history and related mechanisms, but also can hinder the spread of pathogenic microorganisms.
Keywords: Yellow River Wetland; common crane (Grus grus); demoiselle crane (Grus virgo); gut bacteria; gut fungi.
Copyright © 2024 Li, Duan, Wang, Wu, Meng, Bao, Gao and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Differences in Intestinal Microbiota Between White and Common Cranes in the Yellow River Delta During Winter.Biology (Basel). 2025 Jun 16;14(6):704. doi: 10.3390/biology14060704. Biology (Basel). 2025. PMID: 40563956 Free PMC article.
-
Characterization of the gut microbiome of black-necked cranes (Grus nigricollis) in six wintering areas in China.Arch Microbiol. 2020 Jul;202(5):983-993. doi: 10.1007/s00203-019-01802-0. Epub 2020 Jan 4. Arch Microbiol. 2020. PMID: 31901964
-
Comparative analysis of intestinal flora between rare wild red-crowned crane and white-naped crane.Front Microbiol. 2022 Dec 1;13:1007884. doi: 10.3389/fmicb.2022.1007884. eCollection 2022. Front Microbiol. 2022. PMID: 36532425 Free PMC article.
-
Red-crowned Cranes (Grus japonensis) habitat changes in China from 1980 to 2020: Spatio-temporal distribution.J Environ Manage. 2025 Mar;376:124501. doi: 10.1016/j.jenvman.2025.124501. Epub 2025 Feb 15. J Environ Manage. 2025. PMID: 39954500 Review.
-
Winners and losers of land use change: A systematic review of interactions between the world's crane species (Gruidae) and the agricultural sector.Ecol Evol. 2022 Mar 24;12(3):e8719. doi: 10.1002/ece3.8719. eCollection 2022 Mar. Ecol Evol. 2022. PMID: 35356570 Free PMC article. Review.
Cited by
-
Comparative analysis of fecal microbiota of central and eastern black-necked cranes (Grus nigricollis) wintering in Yunnan Province, China.PeerJ. 2025 May 27;13:e19520. doi: 10.7717/peerj.19520. eCollection 2025. PeerJ. 2025. PMID: 40452926 Free PMC article.
-
Analysis of fungal diversity in the feces of Arborophila rufipectus.Front Vet Sci. 2024 Oct 14;11:1430518. doi: 10.3389/fvets.2024.1430518. eCollection 2024. Front Vet Sci. 2024. PMID: 39469585 Free PMC article.
-
Gut fungi of black-necked cranes (Grus nigricollis) respond to dietary changes during wintering.BMC Microbiol. 2024 Jun 29;24(1):232. doi: 10.1186/s12866-024-03396-0. BMC Microbiol. 2024. PMID: 38951807 Free PMC article.
-
Seasonal changes in invertebrate diet of breeding black-necked cranes (Grus nigricollis).Ecol Evol. 2024 Aug 29;14(9):e70234. doi: 10.1002/ece3.70234. eCollection 2024 Sep. Ecol Evol. 2024. PMID: 39219571 Free PMC article.
-
Differences in Intestinal Microbiota Between White and Common Cranes in the Yellow River Delta During Winter.Biology (Basel). 2025 Jun 16;14(6):704. doi: 10.3390/biology14060704. Biology (Basel). 2025. PMID: 40563956 Free PMC article.
References
-
- Ding C. Q., Li F. (2005). Conservation and research of Crested Ibis. Chin. J. Zool. 40, 54–62 (in Chinese).
LinkOut - more resources
Full Text Sources

