Automatic and interpretable prediction of the site of origin in outflow tract ventricular arrhythmias: machine learning integrating electrocardiograms and clinical data
- PMID: 38572307
- PMCID: PMC10987867
- DOI: 10.3389/fcvm.2024.1353096
Automatic and interpretable prediction of the site of origin in outflow tract ventricular arrhythmias: machine learning integrating electrocardiograms and clinical data
Abstract
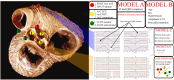
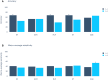
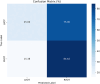
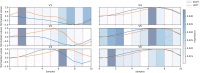
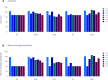
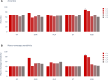
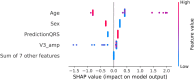
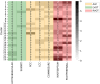
The treatment of outflow tract ventricular arrhythmias (OTVA) through radiofrequency ablation requires the precise identification of the site of origin (SOO). Pinpointing the SOO enhances the likelihood of a successful procedure, reducing intervention times and recurrence rates. Current clinical methods to identify the SOO are based on qualitative analysis of pre-operative electrocardiograms (ECG), heavily relying on physician's expertise. Although computational models and machine learning (ML) approaches have been proposed to assist OTVA procedures, they either consume substantial time, lack interpretability or do not use clinical information. Here, we propose an alternative strategy for automatically predicting the ventricular origin of OTVA patients using ML. Our objective was to classify ventricular (left/right) origin in the outflow tracts (LVOT and RVOT, respectively), integrating ECG and clinical data from each patient. Extending beyond differentiating ventricle origin, we explored specific SOO characterization. Utilizing four databases, we also trained supervised learning models on the QRS complexes of the ECGs, clinical data, and their combinations. The best model achieved an accuracy of 89%, highlighting the significance of precordial leads V1-V4, especially in the R/S transition and initiation of the QRS complex in V2. Unsupervised analysis revealed that some origins tended to group closer than others, e.g., right coronary cusp (RCC) with a less sparse group than the aortic cusp origins, suggesting identifiable patterns for specific SOOs.
Keywords: electrocardiogram; feature analysis; machine learning; outflow tract ventricular arrhythmias; site of origin.
© 2024 Bocanegra-Pérez, Piella, Sebastian, Jimenez-Perez, Falasconi, Saglietto, Soto-Iglesias, Berruezo, Penela and Camara.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

