Rev1 overexpression accelerates N-methyl-N-nitrosourea (MNU)-induced thymic lymphoma by increasing mutagenesis
- PMID: 38572512
- PMCID: PMC11145157
- DOI: 10.1111/cas.16159
Rev1 overexpression accelerates N-methyl-N-nitrosourea (MNU)-induced thymic lymphoma by increasing mutagenesis
Abstract
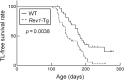
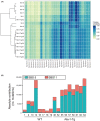
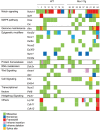
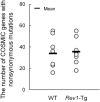
Rev1 has two important functions in the translesion synthesis pathway, including dCMP transferase activity, and acts as a scaffolding protein for other polymerases involved in translesion synthesis. However, the role of Rev1 in mutagenesis and tumorigenesis in vivo remains unclear. We previously generated Rev1-overexpressing (Rev1-Tg) mice and reported that they exhibited a significantly increased incidence of intestinal adenoma and thymic lymphoma (TL) after N-methyl-N-nitrosourea (MNU) treatment. In this study, we investigated mutagenesis of MNU-induced TL tumorigenesis in wild-type (WT) and Rev1-Tg mice using diverse approaches, including whole-exome sequencing (WES). In Rev1-Tg TLs, the mutation frequency was higher than that in WT TL in most cases. However, no difference in the number of nonsynonymous mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC) genes was observed, and mutations involved in Notch1 and MAPK signaling were similarly detected in both TLs. Mutational signature analysis of WT and Rev1-Tg TLs revealed cosine similarity with COSMIC mutational SBS5 (aging-related) and SBS11 (alkylation-related). Interestingly, the total number of mutations, but not the genotypes of WT and Rev1-Tg, was positively correlated with the relative contribution of SBS5 in individual TLs, suggesting that genetic instability could be accelerated in Rev1-Tg TLs. Finally, we demonstrated that preleukemic cells could be detected earlier in Rev1-Tg mice than in WT mice, following MNU treatment. In conclusion, Rev1 overexpression accelerates mutagenesis and increases the incidence of MNU-induced TL by shortening the latency period, which may be associated with more frequent DNA damage-induced genetic instability.
Keywords: DNA translesion synthesis; mutagenesis; thymic lymphoma; tumorigenesis; whole‐exome sequencing.
© 2024 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
YM received the research funds from Amano Enzyme Foundation for Science and Technology and from DAIKO FOUNDATION. These funds have no relevance to the content of this article. Other authors have no conflict of interest.
Figures


References
-
- Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167‐203. - PubMed
-
- Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627‐1630. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317‐353. - PubMed
MeSH terms
Substances
Grants and funding
- Network-Type Joint Usage/Research Center for Radiation Disaster Medical Science at Hiroshima University, Nagasaki University, and Fukushima Medical University
- NIFS10KOBS015/National Institute for Fusion Science Collaborative Research Program
- NIFS13KOBA028/National Institute for Fusion Science Collaborative Research Program
- NIFS20KOCA004/National Institute for Fusion Science Collaborative Research Program
- Initiative for Realizing Diversity in the Research Environment (Specific Correspondence Type), a support project for the Development of Human Resources in Science and Technology conducted by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

