Stable Interstrand Cross-Links Generated from the Repair of 1, N6-Ethenoadenine in DNA by α-Ketoglutarate/Fe(II)-Dependent Dioxygenase ALKBH2
- PMID: 38573229
- PMCID: PMC11060877
- DOI: 10.1021/jacs.3c12890
Stable Interstrand Cross-Links Generated from the Repair of 1, N6-Ethenoadenine in DNA by α-Ketoglutarate/Fe(II)-Dependent Dioxygenase ALKBH2
Abstract
DNA cross-links severely challenge replication and transcription in cells, promoting senescence and cell death. In this paper, we report a novel type of DNA interstrand cross-link (ICL) produced as a side product during the attempted repair of 1,N6-ethenoadenine (εA) by human α-ketoglutarate/Fe(II)-dependent enzyme ALKBH2. This stable/nonreversible ICL was characterized by denaturing polyacrylamide gel electrophoresis analysis and quantified by high-resolution LC-MS in well-matched and mismatched DNA duplexes, yielding 5.7% as the highest level for cross-link formation. The binary lesion is proposed to be generated through covalent bond formation between the epoxide intermediate of εA repair and the exocyclic N6-amino group of adenine or the N4-amino group of cytosine residues in the complementary strand under physiological conditions. The cross-links occur in diverse sequence contexts, and molecular dynamics simulations rationalize the context specificity of cross-link formation. In addition, the cross-link generated from attempted εA repair was detected in cells by highly sensitive LC-MS techniques, giving biological relevance to the cross-link adducts. Overall, a combination of biochemical, computational, and mass spectrometric methods was used to discover and characterize this new type of stable cross-link both in vitro and in human cells, thereby uniquely demonstrating the existence of a potentially harmful ICL during DNA repair by human ALKBH2.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
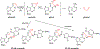
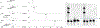




Similar articles
-
Insights into the Direct Oxidative Repair of Etheno Lesions: MD and QM/MM Study on the Substrate Scope of ALKBH2 and AlkB.DNA Repair (Amst). 2020 Dec;96:102944. doi: 10.1016/j.dnarep.2020.102944. Epub 2020 Sep 9. DNA Repair (Amst). 2020. PMID: 33161373 Free PMC article.
-
Differential repair of etheno-DNA adducts by bacterial and human AlkB proteins.DNA Repair (Amst). 2015 Jun;30:1-10. doi: 10.1016/j.dnarep.2015.02.021. Epub 2015 Mar 5. DNA Repair (Amst). 2015. PMID: 25797601 Free PMC article.
-
Comparison of the Base Excision and Direct Reversal Repair Pathways for Correcting 1,N6-Ethenoadenine in Strongly Positioned Nucleosome Core Particles.Chem Res Toxicol. 2020 Jul 20;33(7):1888-1896. doi: 10.1021/acs.chemrestox.0c00089. Epub 2020 May 1. Chem Res Toxicol. 2020. PMID: 32293880 Free PMC article.
-
The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond.J Biol Chem. 2015 Aug 21;290(34):20734-20742. doi: 10.1074/jbc.R115.656462. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152727 Free PMC article. Review.
-
Non-heme dioxygenases in tumor hypoxia: They're all bound with the same fate.DNA Repair (Amst). 2017 Jan;49:21-25. doi: 10.1016/j.dnarep.2016.12.001. Epub 2016 Dec 5. DNA Repair (Amst). 2017. PMID: 27964836 Review.
Cited by
-
Repair of genomic interstrand crosslinks.DNA Repair (Amst). 2024 Sep;141:103739. doi: 10.1016/j.dnarep.2024.103739. Epub 2024 Jul 30. DNA Repair (Amst). 2024. PMID: 39106540 Free PMC article. Review.
References
-
- Stingele J; Bellelli R; Boulton SJ Mechanisms of DNA–protein crosslink repair. Nat. Rev. Mol. Cell Biol 2017, 18, 563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

