Moving Biosurveillance Beyond Coded Data Using AI for Symptom Detection From Physician Notes: Retrospective Cohort Study
- PMID: 38573752
- PMCID: PMC11027052
- DOI: 10.2196/53367
Moving Biosurveillance Beyond Coded Data Using AI for Symptom Detection From Physician Notes: Retrospective Cohort Study
Abstract
Background: Real-time surveillance of emerging infectious diseases necessitates a dynamically evolving, computable case definition, which frequently incorporates symptom-related criteria. For symptom detection, both population health monitoring platforms and research initiatives primarily depend on structured data extracted from electronic health records.
Objective: This study sought to validate and test an artificial intelligence (AI)-based natural language processing (NLP) pipeline for detecting COVID-19 symptoms from physician notes in pediatric patients. We specifically study patients presenting to the emergency department (ED) who can be sentinel cases in an outbreak.
Methods: Subjects in this retrospective cohort study are patients who are 21 years of age and younger, who presented to a pediatric ED at a large academic children's hospital between March 1, 2020, and May 31, 2022. The ED notes for all patients were processed with an NLP pipeline tuned to detect the mention of 11 COVID-19 symptoms based on Centers for Disease Control and Prevention (CDC) criteria. For a gold standard, 3 subject matter experts labeled 226 ED notes and had strong agreement (F1-score=0.986; positive predictive value [PPV]=0.972; and sensitivity=1.0). F1-score, PPV, and sensitivity were used to compare the performance of both NLP and the International Classification of Diseases, 10th Revision (ICD-10) coding to the gold standard chart review. As a formative use case, variations in symptom patterns were measured across SARS-CoV-2 variant eras.
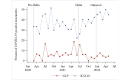
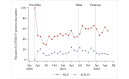
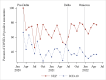
Results: There were 85,678 ED encounters during the study period, including 4% (n=3420) with patients with COVID-19. NLP was more accurate at identifying encounters with patients that had any of the COVID-19 symptoms (F1-score=0.796) than ICD-10 codes (F1-score =0.451). NLP accuracy was higher for positive symptoms (sensitivity=0.930) than ICD-10 (sensitivity=0.300). However, ICD-10 accuracy was higher for negative symptoms (specificity=0.994) than NLP (specificity=0.917). Congestion or runny nose showed the highest accuracy difference (NLP: F1-score=0.828 and ICD-10: F1-score=0.042). For encounters with patients with COVID-19, prevalence estimates of each NLP symptom differed across variant eras. Patients with COVID-19 were more likely to have each NLP symptom detected than patients without this disease. Effect sizes (odds ratios) varied across pandemic eras.
Conclusions: This study establishes the value of AI-based NLP as a highly effective tool for real-time COVID-19 symptom detection in pediatric patients, outperforming traditional ICD-10 methods. It also reveals the evolving nature of symptom prevalence across different virus variants, underscoring the need for dynamic, technology-driven approaches in infectious disease surveillance.
Keywords: AI; COVID-19; SARS-CoV-2; adolescent; adolescents; artificial intelligence; child; children; clinical note; clinical notes; detect; detection; diagnose; diagnosis; diagnostic; diagnostics; documentation; emergency; infectious; natural language processing; paediatric; paediatrics; pediatric; pediatrics; pipeline; pipelines; public health, biosurveillance; pulmonary; respiratory; surveillance; symptom; symptoms; teen; teenager; teenagers; teens; urgent; youth.
©Andrew J McMurry, Amy R Zipursky, Alon Geva, Karen L Olson, James R Jones, Vladimir Ignatov, Timothy A Miller, Kenneth D Mandl. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 04.04.2024.
Conflict of interest statement
Conflicts of Interest: TAM is a member of the advisory council for Lavita AI. Others declare no conflicts of interest.
Figures
References
-
- Subramanian A, Nirantharakumar K, Hughes S, Myles P, Williams T, Gokhale KM, Taverner T, Chandan JS, Brown K, Simms-Williams N, Shah AD, Singh M, Kidy F, Okoth K, Hotham R, Bashir N, Cockburn N, Lee SI, Turner GM, Gkoutos GV, Aiyegbusi OL, McMullan C, Denniston AK, Sapey E, Lord JM, Wraith DC, Leggett E, Iles C, Marshall T, Price MJ, Marwaha S, Davies EH, Jackson LJ, Matthews KL, Camaradou J, Calvert M, Haroon S. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med. 2022;28(8):1706–1714. doi: 10.1038/s41591-022-01909-w. https://europepmc.org/abstract/MED/35879616 10.1038/s41591-022-01909-w - DOI - PMC - PubMed
-
- Crabb BT, Lyons A, Bale M, Martin V, Berger B, Mann S, West WB, Brown A, Peacock JB, Leung DT, Shah RU. Comparison of International Classification of Diseases and Related Health Problems, Tenth Revision codes with electronic medical records among patients with symptoms of coronavirus disease 2019. JAMA Netw Open. 2020;3(8):e2017703. doi: 10.1001/jamanetworkopen.2020.17703. https://europepmc.org/abstract/MED/32797176 2769428 - DOI - PMC - PubMed
-
- Wang J, Abu-El-Rub N, Gray J, Pham HA, Zhou Y, Manion FJ, Liu M, Song X, Xu H, Rouhizadeh M, Zhang Y. COVID-19 SignSym: a fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J Am Med Inform Assoc. 2021;28(6):1275–1283. doi: 10.1093/jamia/ocab015. https://europepmc.org/abstract/MED/33674830 6155732 - DOI - PMC - PubMed
-
- Malden DE, Tartof SY, Ackerson BK, Hong V, Skarbinski J, Yau V, Qian L, Fischer H, Shaw SF, Caparosa S, Xie F. Natural language processing for improved characterization of COVID-19 symptoms: observational study of 350,000 patients in a large integrated health care system. JMIR Public Health Surveill. 2022;8(12):e41529. doi: 10.2196/41529. https://publichealth.jmir.org/2022/12/e41529/ v8i12e41529 - DOI - PMC - PubMed
-
- Di Chiara C, Boracchini R, Sturniolo G, Barbieri A, Costenaro P, Cozzani S, De Pieri M, Liberati C, Zin A, Padoan A, Bonfante F, Kakkar F, Cantarutti A, Donà D, Giaquinto C. Clinical features of COVID-19 in Italian outpatient children and adolescents during parental, Delta, and Omicron waves: a prospective, observational, cohort study. Front Pediatr. 2023;11:1193857. doi: 10.3389/fped.2023.1193857. https://europepmc.org/abstract/MED/37635788 - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

