The Intellectual Disability Risk Gene Kdm5b Regulates Long-Term Memory Consolidation in the Hippocampus
- PMID: 38575342
- PMCID: PMC11079963
- DOI: 10.1523/JNEUROSCI.1544-23.2024
The Intellectual Disability Risk Gene Kdm5b Regulates Long-Term Memory Consolidation in the Hippocampus
Abstract
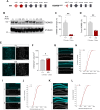
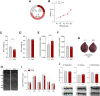
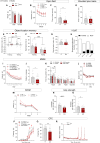
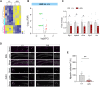
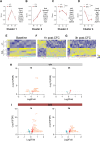
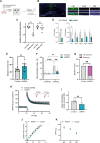
The histone lysine demethylase KDM5B is implicated in recessive intellectual disability disorders, and heterozygous, protein-truncating variants in KDM5B are associated with reduced cognitive function in the population. The KDM5 family of lysine demethylases has developmental and homeostatic functions in the brain, some of which appear to be independent of lysine demethylase activity. To determine the functions of KDM5B in hippocampus-dependent learning and memory, we first studied male and female mice homozygous for a Kdm5b Δ ARID allele that lacks demethylase activity. Kdm5b Δ ARID/ Δ ARID mice exhibited hyperactivity and long-term memory deficits in hippocampus-dependent learning tasks. The expression of immediate early, activity-dependent genes was downregulated in these mice and hyperactivated upon a learning stimulus compared with wild-type (WT) mice. A number of other learning-associated genes were also significantly dysregulated in the Kdm5b Δ ARID/ Δ ARID hippocampus. Next, we knocked down Kdm5b specifically in the adult, WT mouse hippocampus with shRNA. Kdm5b knockdown resulted in spontaneous seizures, hyperactivity, and hippocampus-dependent long-term memory and long-term potentiation deficits. These findings identify KDM5B as a critical regulator of gene expression and synaptic plasticity in the adult hippocampus and suggest that at least some of the cognitive phenotypes associated with KDM5B gene variants are caused by direct effects on memory consolidation mechanisms.
Keywords: KDM5B; chromatin; hippocampus; histone lysine demethylase; learning; memory; mouse.
Copyright © 2024 Perez-Sisques et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K (2013) The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 9:e1003461. 10.1371/journal.pgen.1003461 - DOI - PMC - PubMed
-
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA (2011) Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36:1545–1556. 10.1038/npp.2011.61 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
