The GFPT2-O-GlcNAcylation-YBX1 axis promotes IL-18 secretion to regulate the tumor immune microenvironment in pancreatic cancer
- PMID: 38575607
- PMCID: PMC10995196
- DOI: 10.1038/s41419-024-06589-7
The GFPT2-O-GlcNAcylation-YBX1 axis promotes IL-18 secretion to regulate the tumor immune microenvironment in pancreatic cancer
Abstract
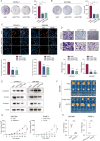
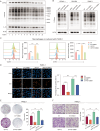
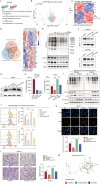
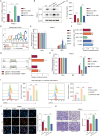
The immunosuppressive microenvironment caused by several intrinsic and extrinsic mechanism has brought great challenges to the immunotherapy of pancreatic cancer. We identified GFPT2, the key enzyme in hexosamine biosynthesis pathway (HBP), as an immune-related prognostic gene in pancreatic cancer using transcriptome sequencing and further confirmed that GFPT2 promoted macrophage M2 polarization and malignant phenotype of pancreatic cancer. HBP is a glucose metabolism pathway leading to the generation of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), which is further utilized for protein O-GlcNAcylation. We confirmed GFPT2-mediated O-GlcNAcylation played an important role in regulating immune microenvironment. Through cellular proteomics, we identified IL-18 as a key downstream of GFPT2 in regulating the immune microenvironment. Through CO-IP and protein mass spectrum, we confirmed that YBX1 was O-GlcNAcylated and nuclear translocated by GFPT2-mediated O-GlcNAcylation. Then, YBX1 functioned as a transcription factor to promote IL-18 transcription. Our study elucidated the relationship between the metabolic pathway of HBP in cancer cells and the immune microenvironment, which might provide some insights into the combination therapy of HBP vulnerability and immunotherapy in pancreatic cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–75. doi: 10.1200/JCO.2009.26.7609. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

