Progeria-based vascular model identifies networks associated with cardiovascular aging and disease
- PMID: 38576084
- PMCID: PMC11258467
- DOI: 10.1111/acel.14150
Progeria-based vascular model identifies networks associated with cardiovascular aging and disease
Abstract
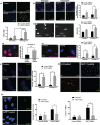
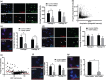
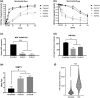
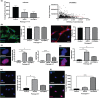
Hutchinson-Gilford Progeria syndrome (HGPS) is a lethal premature aging disorder caused by a de novo heterozygous mutation that leads to the accumulation of a splicing isoform of Lamin A termed progerin. Progerin expression deregulates the organization of the nuclear lamina and the epigenetic landscape. Progerin has also been observed to accumulate at low levels during normal aging in cardiovascular cells of adults that do not carry genetic mutations linked with HGPS. Therefore, the molecular mechanisms that lead to vascular dysfunction in HGPS may also play a role in vascular aging-associated diseases, such as myocardial infarction and stroke. Here, we show that HGPS patient-derived vascular smooth muscle cells (VSMCs) recapitulate HGPS molecular hallmarks. Transcriptional profiling revealed cardiovascular disease remodeling and reactive oxidative stress response activation in HGPS VSMCs. Proteomic analyses identified abnormal acetylation programs in HGPS VSMC replication fork complexes, resulting in reduced H4K16 acetylation. Analysis of acetylation kinetics revealed both upregulation of K16 deacetylation and downregulation of K16 acetylation. This correlates with abnormal accumulation of error-prone nonhomologous end joining (NHEJ) repair proteins on newly replicated chromatin. The knockdown of the histone acetyltransferase MOF recapitulates preferential engagement of NHEJ repair activity in control VSMCs. Additionally, we find that primary donor-derived coronary artery vascular smooth muscle cells from aged individuals show similar defects to HGPS VSMCs, including loss of H4K16 acetylation. Altogether, we provide insight into the molecular mechanisms underlying vascular complications associated with HGPS patients and normative aging.
Keywords: DNA damage repair; H4K16 acetylation; Lamin A; aging; coronary artery; induced pluripotent stem cells; progeria; reprogramming; vascular smooth muscle cells.
© 2024 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
-
- Alagoz, M. , Katsuki, Y. , Ogiwara, H. , Ogi, T. , Shibata, A. , Kakarougkas, A. , & Jeggo, P. (2015). SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Research, 43(16), 7931–7944. 10.1093/nar/gkv722 - DOI - PMC - PubMed
-
- Balmus, G. , Larrieu, D. , Barros, A. C. , Collins, C. , Abrudan, M. , Demir, M. , Geisler, N. J. , Lelliott, C. J. , White, J. K. , Karp, N. A. , Atkinson, J. , Kirton, A. , Jacobsen, M. , Clift, D. , Rodriguez, R. , Sanger Mouse Genetics Project , Shannon, C. , Sanderson, M. , Gates, A. , … Jackson, S. P. (2018). Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nature Communications, 9(1), 1700. 10.1038/s41467-018-03770-3 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

