Transfer learning improves pMHC kinetic stability and immunogenicity predictions
- PMID: 38577265
- PMCID: PMC10994007
- DOI: 10.1016/j.immuno.2023.100030
Transfer learning improves pMHC kinetic stability and immunogenicity predictions
Abstract
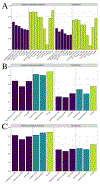
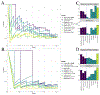
The cellular immune response comprises several processes, with the most notable ones being the binding of the peptide to the Major Histocompability Complex (MHC), the peptide-MHC (pMHC) presentation to the surface of the cell, and the recognition of the pMHC by the T-Cell Receptor. Identifying the most potent peptide targets for MHC binding, presentation and T-cell recognition is vital for developing peptide-based vaccines and T-cell-based immunotherapies. Data-driven tools that predict each of these steps have been developed, and the availability of mass spectrometry (MS) datasets has facilitated the development of accurate Machine Learning (ML) methods for class-I pMHC binding prediction. However, the accuracy of ML-based tools for pMHC kinetic stability prediction and peptide immunogenicity prediction is uncertain, as stability and immunogenicity datasets are not abundant. Here, we use transfer learning techniques to improve stability and immunogenicity predictions, by taking advantage of a large number of binding affinity and MS datasets. The resulting models, TLStab and TLImm, exhibit comparable or better performance than state-of-the-art approaches on different stability and immunogenicity test sets respectively. Our approach demonstrates the promise of learning from the task of peptide binding to improve predictions on downstream tasks. The source code of TLStab and TLImm is publicly available at https://github.com/KavrakiLab/TL-MHC.
Keywords: Machine learning; Peptide immunogenicity; Peptide kinetic stability; Peptide-MHC; Transfer learning.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
NetMHCstab - predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery.Immunology. 2014 Jan;141(1):18-26. doi: 10.1111/imm.12160. Immunology. 2014. PMID: 23927693 Free PMC article.
-
Structural Prediction of Peptide-MHC Binding Modes.Methods Mol Biol. 2022;2405:245-282. doi: 10.1007/978-1-0716-1855-4_13. Methods Mol Biol. 2022. PMID: 35298818 Review.
-
Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity.Eur J Immunol. 2012 Jun;42(6):1405-16. doi: 10.1002/eji.201141774. Eur J Immunol. 2012. PMID: 22678897
-
RankMHC: Learning to Rank Class-I Peptide-MHC Structural Models.J Chem Inf Model. 2024 Dec 9;64(23):8729-8742. doi: 10.1021/acs.jcim.4c01278. Epub 2024 Nov 18. J Chem Inf Model. 2024. PMID: 39555889
-
A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction.Brief Bioinform. 2020 Jul 15;21(4):1119-1135. doi: 10.1093/bib/bbz051. Brief Bioinform. 2020. PMID: 31204427 Free PMC article. Review.
Cited by
-
DiscovEpi: automated whole proteome MHC-I-epitope prediction and visualization.BMC Bioinformatics. 2024 Sep 27;25(1):310. doi: 10.1186/s12859-024-05931-2. BMC Bioinformatics. 2024. PMID: 39333860 Free PMC article.
-
MHC-I-presented non-canonical antigens expand the cancer immunotherapy targets in acute myeloid leukemia.Sci Data. 2024 Aug 1;11(1):831. doi: 10.1038/s41597-024-03660-y. Sci Data. 2024. PMID: 39090129 Free PMC article.
-
Mutations in glioblastoma proteins do not disrupt epitope presentation and recognition, maintaining a specific CD8 T cell immune response potential.Sci Rep. 2024 Jul 19;14(1):16721. doi: 10.1038/s41598-024-67099-2. Sci Rep. 2024. PMID: 39030304 Free PMC article.
References
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular biology of the cell. 4th ed.. New York: Garland: 2002.
-
- Rammensee H-G, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999:50:213–9. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
