Protein oxidation of fucose environments (POFE) reveals fucose-protein interactions
- PMID: 38577366
- PMCID: PMC10988611
- DOI: 10.1039/d3sc06432h
Protein oxidation of fucose environments (POFE) reveals fucose-protein interactions
Abstract
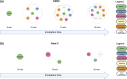
Cell membrane glycoproteins are generally highly fucosylated and sialylated, and post-translational modifications play important roles in the proteins' functions of signaling, binding and cellular processing. For these reasons, methods for measuring sialic acid-mediated protein-protein interactions have been developed. However, determining the role of fucose in these interactions has been limited by technological barriers that have thus far hindered the ability to characterize and observe fucose-mediated protein-protein interactions. Herein, we describe a method to metabolically label mammalian cells with modified fucose, which incorporates a bioorthogonal group into cell membrane glycoproteins thereby enabling the characterization of cell-surface fucose interactome. Copper-catalyzed click chemistry was used to conjugate a proximity labeling probe, azido-FeBABE. Following the addition of hydrogen peroxide (H2O2), the fucose-azido-FeBABE catalyzed the formation of hydroxyl radicals, which in turn oxidized the amino acids in the proximity of the labeled fucose residue. The oxidized peptides were identified using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Variations in degree of protein oxidation were obtained with different H2O2 reaction times yielding the acquisition of spatial information of the fucose-interacting proteins. In addition, specific glycoprotein-protein interactions were constructed for Galectin-3 (LEG3) and Galectin-3-binding protein (LG3BP) illustrating the further utility of the method. This method identifies new fucose binding partners thereby enhancing our understanding of the cell glycocalyx.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
-
- Essentials of Glycobiology, ed. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, D. Mohnen, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 4th edn, 2022 - PubMed

