Soil, rhizosphere, and root microbiome in kiwifruit vine decline, an emerging multifactorial disease
- PMID: 38577679
- PMCID: PMC10991698
- DOI: 10.3389/fmicb.2024.1330865
Soil, rhizosphere, and root microbiome in kiwifruit vine decline, an emerging multifactorial disease
Abstract
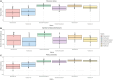
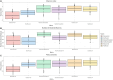
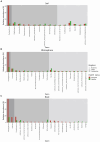
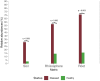
Kiwifruit vine decline syndrome (KVDS) is characterized by severe root system impairment, which leads to irreversible wilting of the canopy. Plants usually collapse rapidly from the appearance of the first aboveground symptoms, without recovery even in the following seasons. The syndrome has been negatively impacting kiwifruit yield in different areas of Italy, the main producing European country, since its first outbreak in 2012. To date, a unique, common causal factor has yet to be found, and the syndrome is referred to as multifactorial. In this article, we investigated the whole biotic community (fungi, bacteria, and oomycetes) associated with the development of KVDS in three different belowground matrices/compartments (soil, rhizosphere, and root). Sampling was performed at both healthy and affected sites located in the main kiwifruit-producing area of Northwestern Italy. To address the multifactorial nature of the syndrome and to investigate the potential roles of abiotic factors in shaping these communities, a physicochemical analysis of soils was also performed. This study investigates the associations among taxonomic groups composing the microbiome and also between biotic and abiotic factors. Dysbiosis was considered as a driving event in shaping KVDS microbial communities. The results obtained from this study highlight the role of the oomycete genus Phytopythium, which resulted predominantly in the oomycete community composition of diseased matrices, though it was also present in healthy ones. Both bacterial and fungal communities resulted in a high richness of genera and were highly correlated to the sampling site and matrix, underlining the importance of multiple location sampling both geographically and spatially. The rhizosphere community associated with KVDS was driven by a dysbiotic process. In addition, analysis of the association network in the diseased rhizosphere revealed the presence of potential cross-kingdom competition for plant-derived carbon between saprobes, oomycetes, and bacteria.
Keywords: Phytopythium; dysbiosis; kiwifruit vine decline syndrome; metabarcoding; microbiome; multifactorial disease; next-generation sequencing.
Copyright © 2024 Guaschino, Garello, Nari, Zhimo, Droby and Spadaro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Abarenkov K., Zirk A., Piirmann T., Pöhönen R., Ivanov F., Nilsson R. H., et al. (2021). UNITE QIIME release for eukaryotes 2. UNITE Commun. doi: 10.15156/BIO/126486 - DOI
-
- Adamo I., Castaño C., Bonet J. A., Colinas C., Martínez de Aragón J., Alday J. G. (2021). Soil physico-chemical properties have a greater effect on soil fungi than host species in Mediterranean pure and mixed pine forests. Soil Biol. Biochem. 160:108320. doi: 10.1016/j.soilbio.2021.108320 - DOI
-
- Akilli S., Serçe Çi̇ U., Zekaİ Katircioğlu Y., Karakaya A., Maden S. (2011). Involvement of Phytophthora citrophthora in kiwifruit decline in Turkey: involvement of Phytophthora citrophthora on kiwifruit. J. Phytopathol. 159, 579–581. doi: 10.1111/j.1439-0434.2011.01798.x - DOI
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance: non-parametric MANOVA for ecology. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x - DOI
LinkOut - more resources
Full Text Sources

