Sse1, Hsp110 chaperone of yeast, controls the cellular fate during endoplasmic reticulum stress
- PMID: 38577891
- PMCID: PMC11152076
- DOI: 10.1093/g3journal/jkae075
Sse1, Hsp110 chaperone of yeast, controls the cellular fate during endoplasmic reticulum stress
Abstract
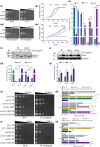
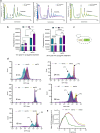
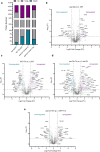
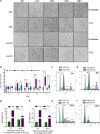
Sse1 is a cytosolic Hsp110 molecular chaperone of yeast, Saccharomyces cerevisiae. Its multifaceted roles in cellular protein homeostasis as a nucleotide exchange factor (NEF), as a protein-disaggregase and as a chaperone linked to protein synthesis (CLIPS) are well documented. In the current study, we show that SSE1 genetically interacts with IRE1 and HAC1, the endoplasmic reticulum-unfolded protein response (ER-UPR) sensors implicating its role in ER protein homeostasis. Interestingly, the absence of this chaperone imparts unusual resistance to tunicamycin-induced ER stress which depends on the intact Ire1-Hac1 mediated ER-UPR signaling. Furthermore, cells lacking SSE1 show inefficient ER-stress-responsive reorganization of translating ribosomes from polysomes to monosomes that drive uninterrupted protein translation during tunicamycin stress. In consequence, the sse1Δ strain shows prominently faster reversal from ER-UPR activated state indicating quicker restoration of homeostasis, in comparison to the wild-type (WT) cells. Importantly, Sse1 plays a critical role in controlling the ER-stress-mediated cell division arrest, which is escaped in sse1Δ strain during chronic tunicamycin stress. Accordingly, sse1Δ strain shows significantly higher cell viability in comparison to WT yeast imparting the stark fitness following short-term as well as long-term tunicamycin stress. These data, all together, suggest that cytosolic chaperone Sse1 is an important modulator of ER stress response in yeast and it controls stress-induced cell division arrest and cell death during overwhelming ER stress induced by tunicamycin.
Keywords: endoplasmic reticulum stress; endoplasmic reticulum unfolded protein response; heat shock protein 110; molecular chaperone; protein homeostasis.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures



References
-
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151(5):1042–1054. doi:10.1016/j.cell.2012.10.044. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
