PaCS-Toolkit: Optimized Software Utilities for Parallel Cascade Selection Molecular Dynamics (PaCS-MD) Simulations and Subsequent Analyses
- PMID: 38578072
- PMCID: PMC11033871
- DOI: 10.1021/acs.jpcb.4c01271
PaCS-Toolkit: Optimized Software Utilities for Parallel Cascade Selection Molecular Dynamics (PaCS-MD) Simulations and Subsequent Analyses
Abstract
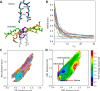
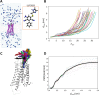
Parallel cascade selection molecular dynamics (PaCS-MD) is an enhanced conformational sampling method conducted as a "repetition of time leaps in parallel worlds", comprising cycles of multiple molecular dynamics (MD) simulations performed in parallel and selection of the initial structures of MDs for the next cycle. We developed PaCS-Toolkit, an optimized software utility enabling the use of different MD software and trajectory analysis tools to facilitate the execution of the PaCS-MD simulation and analyze the obtained trajectories, including the preparation for the subsequent construction of the Markov state model. PaCS-Toolkit is coded with Python, is compatible with various computing environments, and allows for easy customization by editing the configuration file and specifying the MD software and analysis tools to be used. We present the software design of PaCS-Toolkit and demonstrate applications of PaCS-MD variations: original targeted PaCS-MD to peptide folding; rmsdPaCS-MD to protein domain motion; and dissociation PaCS-MD to ligand dissociation from adenosine A2A receptor.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Nontargeted Parallel Cascade Selection Molecular Dynamics for Enhancing the Conformational Sampling of Proteins.J Chem Theory Comput. 2015 Nov 10;11(11):5493-502. doi: 10.1021/acs.jctc.5b00723. Epub 2015 Oct 21. J Chem Theory Comput. 2015. PMID: 26574337
-
Nontargeted Parallel Cascade Selection Molecular Dynamics Based on a Nonredundant Selection Rule for Initial Structures Enhances Conformational Sampling of Proteins.J Chem Inf Model. 2019 Dec 23;59(12):5198-5206. doi: 10.1021/acs.jcim.9b00753. Epub 2019 Nov 21. J Chem Inf Model. 2019. PMID: 31697897
-
Molecular Simulation to Investigate Open-Close Motion of a Flagellar Export Apparatus Protein FlhAC.Methods Mol Biol. 2023;2646:27-34. doi: 10.1007/978-1-0716-3060-0_3. Methods Mol Biol. 2023. PMID: 36842103
-
How to Run FAST Simulations.Methods Enzymol. 2016;578:213-25. doi: 10.1016/bs.mie.2016.05.032. Epub 2016 Jun 16. Methods Enzymol. 2016. PMID: 27497168 Review.
-
Detecting Functional Dynamics in Proteins with Comparative Perturbed-Ensembles Analysis.Acc Chem Res. 2019 Dec 17;52(12):3455-3464. doi: 10.1021/acs.accounts.9b00485. Epub 2019 Dec 3. Acc Chem Res. 2019. PMID: 31793290 Review.
Cited by
-
Product release and substrate entry of aldehyde deformylating oxygenase revealed by molecular dynamics simulations.Biophys Physicobiol. 2025 Jan 9;22(1):e220003. doi: 10.2142/biophysico.bppb-v22.0003. eCollection 2025. Biophys Physicobiol. 2025. PMID: 40046558 Free PMC article.
-
RNA Binding Mechanism of the FUS Zinc Finger in Concert with Its Flanking Intrinsically Disordered Region.J Chem Inf Model. 2025 Aug 11;65(15):8262-8275. doi: 10.1021/acs.jcim.5c01059. Epub 2025 Jul 22. J Chem Inf Model. 2025. PMID: 40693919 Free PMC article.
-
Balancing G protein selectivity and efficacy in the adenosine A2A receptor.Nat Chem Biol. 2025 Jan;21(1):71-79. doi: 10.1038/s41589-024-01682-6. Epub 2024 Jul 31. Nat Chem Biol. 2025. PMID: 39085516
-
Unveiling the antiviral inhibitory activity of ebselen and ebsulfur derivatives on SARS-CoV-2 using machine learning-based QSAR, LB-PaCS-MD, and experimental assay.Sci Rep. 2025 Feb 26;15(1):6956. doi: 10.1038/s41598-025-91235-1. Sci Rep. 2025. PMID: 40011571 Free PMC article.
-
Structural Basis for the Catalytic Mechanism of ATP-Dependent Diazotase CmaA6.Angew Chem Int Ed Engl. 2025 Jul;64(27):e202505851. doi: 10.1002/anie.202505851. Epub 2025 May 5. Angew Chem Int Ed Engl. 2025. PMID: 40275441 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

