Genetic diversity and phylogeographic patterns of the peacock jewel-damselfly, Rhinocypha fenestrella (Rambur, 1842)
- PMID: 38578719
- PMCID: PMC10997100
- DOI: 10.1371/journal.pone.0301392
Genetic diversity and phylogeographic patterns of the peacock jewel-damselfly, Rhinocypha fenestrella (Rambur, 1842)
Abstract
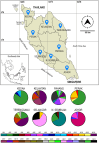
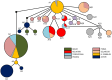
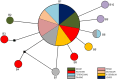
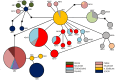
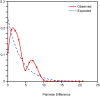
Despite is known to have widespread distribution and the most active species of the family Chlorocyphidae, the molecular data of Rhinocypha fenestrella (Rambur, 1842) are relatively scarce. The present study is the first that examined the genetic diversity and phylogeographic pattern of the peacock jewel-damselfly R. fenestrella by sequencing the cytochrome C oxidase I (cox1) and 16S rRNA gene regions from 147 individuals representing eight populations in Malaysia. A total of 26 and 10 unique haplotypes were revealed by the cox1 and 16S rRNA genes, respectively, and 32 haplotypes were recovered by the concatenated sequences of cox1+16S. Analyses indicated that haplotype AB2 was the most frequent and the most widespread haplotype in Malaysia while haplotype AB1 was suggested as the common ancestor haplotype of the R. fenestrella that may arose from the Negeri Sembilan as discovered from cox1+16S haplotype network analysis. Overall haplotype and nucleotide diversities of the concatenated sequences were Hd = 0.8937 and Pi = 0.0028, respectively, with great genetic differentiation (FST = 0.6387) and low gene flow (Nm = 0.14). Population from Pahang presented the highest genetic diversity (Hd = 0.8889, Pi = 0.0022, Nh = 9), whereas Kedah population demonstrated the lowest diversity (Hd = 0.2842, Pi = 0.0003, Nh = 4). The concatenated sequences of cox1+16S showed genetic divergence ranging from 0.09% to 0.97%, whereas the genetic divergence for cox1 and 16S rRNA genes were 0.16% to 1.63% and 0.01% to 0.75% respectively. This study provides for the first-time insights on the intraspecific genetic diversity, phylogeographic pattern and ancestral haplotype of Rhinocypha fenestrella. The understanding of molecular data especially phylogeographic pattern can enhance the knowledge about insect origin, their diversity, and capability to disperse in particular environments.
Copyright: © 2024 Noorhidayah et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Lieftinck MA. Two interesting new insular Rhinocypha from Malaysia (Odon.). Tijdschrift Voor Entomologie. 1947; 88: 215–224.
-
- Introduction Córdoba–Aguilar A. In: Córdoba–Aguilar A, editor. Dragonflies and damselflies: study models in ecological and evolutionary research. Oxford (UK): Oxford University Press; 2008. p. 1–3.
-
- Sharma G. Rhinocypha fenestrella. The IUCN Red List of Threatened Species. 2010.
-
- Kahilainen A, Puurtinen M, Kotiaho JS. Conservation implications of species–genetic diversity correlations. Global Ecology and Conservation. 2014; 2: 315–323.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

