Belt and braces: Two escape ways to maintain the cassette reservoir of large chromosomal integrons
- PMID: 38578806
- PMCID: PMC11023631
- DOI: 10.1371/journal.pgen.1011231
Belt and braces: Two escape ways to maintain the cassette reservoir of large chromosomal integrons
Abstract
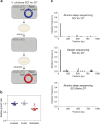
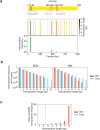
Integrons are adaptive devices that capture, stockpile, shuffle and express gene cassettes thereby sampling combinatorial phenotypic diversity. Some integrons called sedentary chromosomal integrons (SCIs) can be massive structures containing hundreds of cassettes. Since most of these cassettes are non-expressed, it is not clear how they remain stable over long evolutionary timescales. Recently, it was found that the experimental inversion of the SCI of Vibrio cholerae led to a dramatic increase of the cassette excision rate associated with a fitness defect. Here, we question the evolutionary sustainability of this apparently counter selected genetic context. Through experimental evolution, we find that the integrase is rapidly inactivated and that the inverted SCI can recover its original orientation by homologous recombination between two insertion sequences (ISs) present in the array. These two outcomes of SCI inversion restore the normal growth and prevent the loss of cassettes, enabling SCIs to retain their roles as reservoirs of functions. These results illustrate a nice interplay between gene orientation, genome rearrangement, bacterial fitness and demonstrate how integrons can benefit from their embedded ISs.
Copyright: © 2024 Richard et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

