The genetic regulatory architecture and epigenomic basis for age-related changes in rattlesnake venom
- PMID: 38578985
- PMCID: PMC11032440
- DOI: 10.1073/pnas.2313440121
The genetic regulatory architecture and epigenomic basis for age-related changes in rattlesnake venom
Erratum in
-
Correction for Hogan et al., The genetic regulatory architecture and epigenomic basis for age-related changes in rattlesnake venom.Proc Natl Acad Sci U S A. 2025 Mar 18;122(11):e2502598122. doi: 10.1073/pnas.2502598122. Epub 2025 Feb 28. Proc Natl Acad Sci U S A. 2025. PMID: 40020184 Free PMC article. No abstract available.
Abstract
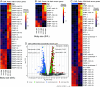
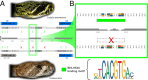
Developmental phenotypic changes can evolve under selection imposed by age- and size-related ecological differences. Many of these changes occur through programmed alterations to gene expression patterns, but the molecular mechanisms and gene-regulatory networks underlying these adaptive changes remain poorly understood. Many venomous snakes, including the eastern diamondback rattlesnake (Crotalus adamanteus), undergo correlated changes in diet and venom expression as snakes grow larger with age, providing models for identifying mechanisms of timed expression changes that underlie adaptive life history traits. By combining a highly contiguous, chromosome-level genome assembly with measures of expression, chromatin accessibility, and histone modifications, we identified cis-regulatory elements and trans-regulatory factors controlling venom ontogeny in the venom glands of C. adamanteus. Ontogenetic expression changes were significantly correlated with epigenomic changes within genes, immediately adjacent to genes (e.g., promoters), and more distant from genes (e.g., enhancers). We identified 37 candidate transcription factors (TFs), with the vast majority being up-regulated in adults. The ontogenetic change is largely driven by an increase in the expression of TFs associated with growth signaling, transcriptional activation, and circadian rhythm/biological timing systems in adults with corresponding epigenomic changes near the differentially expressed venom genes. However, both expression activation and repression contributed to the composition of both adult and juvenile venoms, demonstrating the complexity and potential evolvability of gene regulation for this trait. Overall, given that age-based trait variation is common across the tree of life, we provide a framework for understanding gene-regulatory-network-driven life-history evolution more broadly.
Keywords: Crotalus; epigenomics; gene regulatory network; ontogeny; venom.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




Comment in
-
Coming of age in venom research.Proc Natl Acad Sci U S A. 2024 May 7;121(19):e2405708121. doi: 10.1073/pnas.2405708121. Epub 2024 Apr 30. Proc Natl Acad Sci U S A. 2024. PMID: 38687800 Free PMC article. No abstract available.
References
-
- Erwin D. H., Davidson E. H., The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 10, 141–148 (2009). - PubMed
-
- Hoekstra H. E., Coyne J. A., The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007). - PubMed
-
- Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

