Multi-omics analysis reveals that linoleic acid metabolism is associated with variations of trained immunity induced by distinct BCG strains
- PMID: 38578989
- PMCID: PMC10997199
- DOI: 10.1126/sciadv.adk8093
Multi-omics analysis reveals that linoleic acid metabolism is associated with variations of trained immunity induced by distinct BCG strains
Abstract
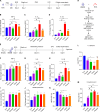
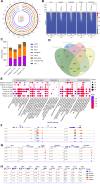
Trained immunity is one of the mechanisms by which BCG vaccination confers persistent nonspecific protection against diverse diseases. Genomic differences between the different BCG vaccine strains that are in global use could result in variable protection against tuberculosis and therapeutic effects on bladder cancer. In this study, we found that four representative BCG strains (BCG-Russia, BCG-Sweden, BCG-China, and BCG-Pasteur) covering all four genetic clusters differed in their ability to induce trained immunity and nonspecific protection. The trained immunity induced by BCG was associated with the Akt-mTOR-HIF1α axis, glycolysis, and NOD-like receptor signaling pathway. Multi-omics analysis (epigenomics, transcriptomics, and metabolomics) showed that linoleic acid metabolism was correlated with the trained immunity-inducing capacity of different BCG strains. Linoleic acid participated in the induction of trained immunity and could act as adjuvants to enhance BCG-induced trained immunity, revealing a trained immunity-inducing signaling pathway that could be used in the adjuvant development.
Figures






References
-
- Fanucchi S., Domínguez-Andrés J., Joosten L. A. B., Netea M. G., Mhlanga M. M., The intersection of epigenetics and metabolism in trained immunity. Immunity 54, 32–43 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

