Cellular atlas of the human ovary using morphologically guided spatial transcriptomics and single-cell sequencing
- PMID: 38578993
- PMCID: PMC10997207
- DOI: 10.1126/sciadv.adm7506
Cellular atlas of the human ovary using morphologically guided spatial transcriptomics and single-cell sequencing
Abstract
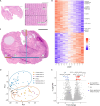
The reproductive and endocrine functions of the ovary involve spatially defined interactions among specialized cell populations. Despite the ovary's importance in fertility and endocrine health, functional attributes of ovarian cells are largely uncharacterized. Here, we profiled >18,000 genes in 257 regions from the ovaries of two premenopausal donors to examine the functional units in the ovary. We also generated single-cell RNA sequencing data for 21,198 cells from three additional donors and identified four major cell types and four immune cell subtypes. Custom selection of sampling areas revealed distinct gene activities for oocytes, theca, and granulosa cells. These data contributed panels of oocyte-, theca-, and granulosa-specific genes, thus expanding the knowledge of molecular programs driving follicle development. Serial samples around oocytes and across the cortex and medulla uncovered previously unappreciated variation of hormone and extracellular matrix remodeling activities. This combined spatial and single-cell atlas serves as a resource for future studies of rare cells and pathological states in the ovary.
Figures




References
-
- K. Hummitzsch, H. F. Irving-Rodgers, J. Schwartz, R. J. Rodgers, Development of the mammalian ovary and follicles, in The Ovary (Elsevier, 2019), pp. 71–82.
-
- O’Neill K. E., Maher J. Y., Laronda M. M., Duncan F. E., LeDuc R. D., Lujan M. E., Oktay K. H., Pouch A. M., Segars J. H., Tsui E. L., Zelinski M. B., Halvorson L. M., Gomez-Lobo V., Anatomic nomenclature and 3-dimensional regional model of the human ovary: Call for a new paradigm. Am. J. Obstet. Gynecol. 228, 270–275.e4 (2023). - PMC - PubMed
-
- D. L. Russell, R. L. Robker, Ovulation: The coordination of intrafollicular networks to ensure oocyte release, in The Ovary (Elsevier, 2019), pp. 217–234.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

