A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5- and CXCR4-tropic HIV-1 infection
- PMID: 38579682
- PMCID: PMC11212398
- DOI: 10.1016/j.stem.2024.03.002
A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5- and CXCR4-tropic HIV-1 infection
Abstract
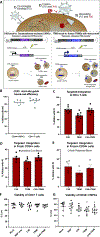
Allogeneic hematopoietic stem and progenitor cell transplant (HSCT) of CCR5 null (CCR5Δ32) cells can be curative for HIV-1-infected patients. However, because allogeneic HSCT poses significant risk, CCR5Δ32 matched bone marrow donors are rare, and CCR5Δ32 transplant does not confer resistance to the CXCR4-tropic virus, it is not a viable option for most patients. We describe a targeted Cas9/AAV6-based genome editing strategy for autologous HSCT resulting in both CCR5- and CXCR4-tropic HIV-1 resistance. Edited human hematopoietic stem and progenitor cells (HSPCs) maintain multi-lineage repopulation capacity in vivo, and edited primary human T cells potently inhibit infection by both CCR5-tropic and CXCR4-tropic HIV-1. Modification rates facilitated complete loss of CCR5-tropic replication and up to a 2,000-fold decrease in CXCR4-tropic replication without CXCR4 locus disruption. This multi-factor editing strategy in HSPCs could provide a broad approach for autologous HSCT as a functional cure for both CCR5-tropic and CXCR4-tropic HIV-1 infections.
Keywords: CCR5 knockout; CRISPR-Cas9; HIV; HIV restriction; autologous HSCT; cell therapy; functional cure; gene editing; hematopoietic stem cell transplant; homology-directed repair.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests This work was supported by the Laurie Kraus Lacob Faculty Scholar Fund in Pediatric Translational Medicine and the Sutardja Chuk Professorship in Definitive and Curative Medicine. M.H.P. has equity in CRISPR Tx, Graphite Bio, Allogene Tx, and Kamau Tx and serves on the SAB for Allogene Tx and on the Board of Directors for Graphite Bio and Kamau Tx. M.H.P. is a Board Observer at Arbor Tx. The authors can confirm that all relevant data are included in the article and/or its supplemental information files or are available upon request.
Figures






References
-
- Kemnic TR, and Gulick PG (2022). HIV Antiretroviral Therapy. In StatPearls, (StatPearls Publishing; Copyright © 2022, StatPearls Publishing LLC.). - PubMed
-
- UNAIDS epidemiological estimates. (2021).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

