Channeling Nicotinamide Phosphoribosyltransferase (NAMPT) to Address Life and Death
- PMID: 38580317
- PMCID: PMC11056997
- DOI: 10.1021/acs.jmedchem.3c02112
Channeling Nicotinamide Phosphoribosyltransferase (NAMPT) to Address Life and Death
Abstract
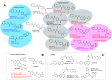
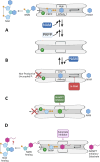
Nicotinamide phosphoribosyltransferase (NAMPT) catalyzes the rate-limiting step in NAD+ biosynthesis via salvage of NAM formed from catabolism of NAD+ by proteins with NADase activity (e.g., PARPs, SIRTs, CD38). Depletion of NAD+ in aging, neurodegeneration, and metabolic disorders is addressed by NAD+ supplementation. Conversely, NAMPT inhibitors have been developed for cancer therapy: many discovered by phenotypic screening for cancer cell death have low nanomolar potency in cellular models. No NAMPT inhibitor is yet FDA-approved. The ability of inhibitors to act as NAMPT substrates may be associated with efficacy and toxicity. Some 3-pyridyl inhibitors become 4-pyridyl activators or "NAD+ boosters". NAMPT positive allosteric modulators (N-PAMs) and boosters may increase enzyme activity by relieving substrate/product inhibition. Binding to a "rear channel" extending from the NAMPT active site is key for inhibitors, boosters, and N-PAMs. A deeper understanding may fulfill the potential of NAMPT ligands to regulate cellular life and death.
Conflict of interest statement
The authors declare the following competing financial interest(s): G.R.J.T. is an inventor on patents assigned to the University of Illinois.
Figures





References
-
- Luongo T. S.; Eller J. M.; Lu M. J.; Niere M.; Raith F.; Perry C.; Bornstein M. R.; Oliphint P.; Wang L.; McReynolds M. R.; Migaud M. E.; Rabinowitz J. D.; Johnson F. B.; Johnsson K.; Ziegler M.; Cambronne X. A.; Baur J. A. SLC25A51 is a mammalian mitochondrial NAD(+) transporter. Nature 2020, 588, 174–179. 10.1038/s41586-020-2741-7. - DOI - PMC - PubMed
-
- Girardi E.; Agrimi G.; Goldmann U.; Fiume G.; Lindinger S.; Sedlyarov V.; Srndic I.; Gurtl B.; Agerer B.; Kartnig F.; Scarcia P.; Di Noia M. A.; Lineiro E.; Rebsamen M.; Wiedmer T.; Bergthaler A.; Palmieri L.; Superti-Furga G. Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nat. Commun. 2020, 11, 6145.10.1038/s41467-020-19871-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

