Cryo-EM structures of pannexin 1 and 3 reveal differences among pannexin isoforms
- PMID: 38580658
- PMCID: PMC10997603
- DOI: 10.1038/s41467-024-47142-6
Cryo-EM structures of pannexin 1 and 3 reveal differences among pannexin isoforms
Abstract
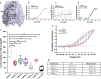
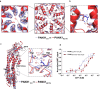
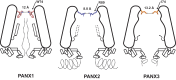
Pannexins are single-membrane large-pore channels that release ions and ATP upon activation. Three isoforms of pannexins 1, 2, and 3, perform diverse cellular roles and differ in their pore lining residues. In this study, we report the cryo-EM structure of pannexin 3 at 3.9 Å and analyze its structural differences with pannexin isoforms 1 and 2. The pannexin 3 vestibule has two distinct chambers and a wider pore radius in comparison to pannexins 1 and 2. We further report two cryo-EM structures of pannexin 1, with pore substitutions W74R/R75D that mimic the pore lining residues of pannexin 2 and a germline mutant of pannexin 1, R217H at resolutions of 3.2 Å and 3.9 Å, respectively. Substitution of cationic residues in the vestibule of pannexin 1 results in reduced ATP interaction propensities to the channel. The germline mutant R217H in transmembrane helix 3 (TM3), leads to a partially constricted pore, reduced ATP interaction and weakened voltage sensitivity. The study compares the three pannexin isoform structures, the effects of substitutions of pore and vestibule-lining residues and allosteric effects of a pathological substitution on channel structure and function thereby enhancing our understanding of this vital group of ATP-release channels.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

