Gene trajectory inference for single-cell data by optimal transport metrics
- PMID: 38580861
- PMCID: PMC11452571
- DOI: 10.1038/s41587-024-02186-3
Gene trajectory inference for single-cell data by optimal transport metrics
Abstract
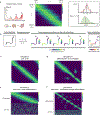
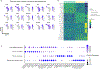
Single-cell RNA sequencing has been widely used to investigate cell state transitions and gene dynamics of biological processes. Current strategies to infer the sequential dynamics of genes in a process typically rely on constructing cell pseudotime through cell trajectory inference. However, the presence of concurrent gene processes in the same group of cells and technical noise can obscure the true progression of the processes studied. To address this challenge, we present GeneTrajectory, an approach that identifies trajectories of genes rather than trajectories of cells. Specifically, optimal transport distances are calculated between gene distributions across the cell-cell graph to extract gene programs and define their gene pseudotemporal order. Here we demonstrate that GeneTrajectory accurately extracts progressive gene dynamics in myeloid lineage maturation. Moreover, we show that GeneTrajectory deconvolves key gene programs underlying mouse skin hair follicle dermal condensate differentiation that could not be resolved by cell trajectory approaches. GeneTrajectory facilitates the discovery of gene programs that control the changes and activities of biological processes.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: R.A.F. is an advisor to GlaxoSmithKline, Zai Lab and Ventus Therapeutics. F.S. is employed as a director by PCMGF Limited. I.D.O. is the founder and president of Plythera and receives research funding from Ventus Therapeutics and SenTry.
Figures
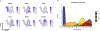








Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
-
OneSC: a computational platform for recapitulating cell state transitions.Bioinformatics. 2024 Nov 28;40(12):btae703. doi: 10.1093/bioinformatics/btae703. Bioinformatics. 2024. PMID: 39570626 Free PMC article.
-
scTsI: an effective two-stage imputation method for single-cell RNA-seq data.Brief Bioinform. 2025 May 1;26(3):bbaf298. doi: 10.1093/bib/bbaf298. Brief Bioinform. 2025. PMID: 40579791 Free PMC article.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
Cited by
-
Exploring structured molecular landscape from single-cell multi-omics data by an explainable multimodal model.iScience. 2024 Nov 2;27(12):111131. doi: 10.1016/j.isci.2024.111131. eCollection 2024 Dec 20. iScience. 2024. PMID: 39618495 Free PMC article.
-
The spatial landscape of glial pathology and T cell response in Parkinson's disease substantia nigra.Nat Commun. 2025 Aug 4;16(1):7146. doi: 10.1038/s41467-025-62478-3. Nat Commun. 2025. PMID: 40759663 Free PMC article.
-
Cilta-cel salvages ide-cel failure in relapsed multiple myeloma by driving distinct immune responses.medRxiv [Preprint]. 2025 Jul 11:2025.07.10.25331322. doi: 10.1101/2025.07.10.25331322. medRxiv. 2025. PMID: 40672475 Free PMC article. Preprint.
-
Investigation of cell development and tissue structure network based on natural Language processing of scRNA-seq data.J Transl Med. 2025 Mar 4;23(1):264. doi: 10.1186/s12967-025-06263-2. J Transl Med. 2025. PMID: 40038714 Free PMC article.
-
Single-cell meta-analysis of T cells reveals clonal dynamics of response to checkpoint immunotherapy.Cell Genom. 2025 May 14;5(5):100842. doi: 10.1016/j.xgen.2025.100842. Epub 2025 Apr 4. Cell Genom. 2025. PMID: 40187353 Free PMC article.
References
Methods-only references
-
- Balasubramanian Mukund and Schwartz Eric L. The isomap algorithm and topological stability. Science, 295(5552):7–7, 2002. - PubMed
-
- Bernstein Mira, De Silva Vin, Langford John C, and Tenenbaum Joshua B. Graph approximations to geodesics on embedded manifolds. Technical report, Department of Psychology, Stanford University, 2000.
-
- Dassule Hèlène R, Lewis Paula, Bei Marianna, Maas Richard, and McMahon Andrew P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development, 127(22):4775–4785, 2000. - PubMed
MeSH terms
Grants and funding
- R01AR076420/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- UM1 DA051410/DA/NIDA NIH HHS/United States
- K08 AR077689/AR/NIAMS NIH HHS/United States
- UM1DA051410/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- U01 DA053628/DA/NIDA NIH HHS/United States
- R01GM131642/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- P50 CA121974/CA/NCI NIH HHS/United States
- R01 GM131642/GM/NIGMS NIH HHS/United States
- U54 AG079759/AG/NIA NIH HHS/United States
- DMS-223784/NSF | Directorate for Mathematical & Physical Sciences | Division of Mathematical Sciences (DMS)
- R03 AR083454/AR/NIAMS NIH HHS/United States
- R01 AI141009/AI/NIAID NIH HHS/United States
- U54AG079759/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- U01DA053628/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- R01 AR076420/AR/NIAMS NIH HHS/United States
- U54 AG076043/AG/NIA NIH HHS/United States
- P50CA121974/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U54AG076043/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
LinkOut - more resources
Full Text Sources

