Apiospora arundinis, a panoply of carbohydrate-active enzymes and secondary metabolites
- PMID: 38582937
- PMCID: PMC10999098
- DOI: 10.1186/s43008-024-00141-0
Apiospora arundinis, a panoply of carbohydrate-active enzymes and secondary metabolites
Abstract
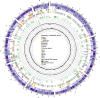
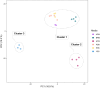
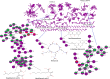
The Apiospora genus comprises filamentous fungi with promising potential, though its full capabilities remain undiscovered. In this study, we present the first genome assembly of an Apiospora arundinis isolate, demonstrating a highly complete and contiguous assembly estimated to 48.8 Mb, with an N99 of 3.0 Mb. Our analysis predicted a total of 15,725 genes, with functional annotations for 13,619 of them, revealing a fungus capable of producing very high amounts of carbohydrate-active enzymes (CAZymes) and secondary metabolites. Through transcriptomic analysis, we observed differential gene expression in response to varying growth media, with several genes related to carbohydrate metabolism showing significant upregulation when the fungus was cultivated on a hay-based medium. Finally, our metabolomic analysis unveiled a fungus capable of producing a diverse array of metabolites.
Keywords: Apiospora; Arthrinium; CAZymes; Oxford Nanopore sequencing; Secondary metabolites.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Bagherabadi S, Zafari D, Ghobadi Anvar F. First report of leaf spot caused by Arthrinium arundinis on rosemary in Iran. J Plant Pathol. 2014;96(S4):126. doi: 10.4454/JPP.V96I4.017. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
