Mesenchymal-specific Alms1 knockout in mice recapitulates metabolic features of Alström syndrome
- PMID: 38583571
- PMCID: PMC11047791
- DOI: 10.1016/j.molmet.2024.101933
Mesenchymal-specific Alms1 knockout in mice recapitulates metabolic features of Alström syndrome
Abstract
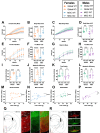
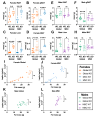
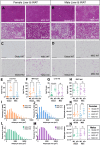
Objective: Alström Syndrome (AS), caused by biallelic ALMS1 mutations, includes obesity with disproportionately severe insulin resistant diabetes, dyslipidemia, and fatty liver. Prior studies suggest that hyperphagia is accounted for by loss of ALMS1 function in hypothalamic neurones, whereas disproportionate metabolic complications may be due to impaired adipose tissue expandability. We tested this by comparing the metabolic effects of global and mesenchymal stem cell (MSC)-specific Alms1 knockout.
Methods: Global Alms1 knockout (KO) mice were generated by crossing floxed Alms1 and CAG-Cre mice. A Pdgfrα-Cre driver was used to abrogate Alms1 function selectively in MSCs and their descendants, including preadipocytes. We combined metabolic phenotyping of global and Pdgfrα+ Alms1-KO mice on a 45% fat diet with measurements of body composition and food intake, and histological analysis of metabolic tissues.
Results: Assessed on 45% fat diet to promote adipose expansion, global Alms1 KO caused hyperphagia, obesity, insulin resistance, dyslipidaemia, and fatty liver. Pdgfrα-cre driven KO of Alms1 (MSC KO) recapitulated insulin resistance, fatty liver, and dyslipidaemia in both sexes. Other phenotypes were sexually dimorphic: increased fat mass was only present in female Alms1 MSC KO mice. Hyperphagia was not evident in male Alms1 MSC KO mice, but was found in MSC KO females, despite no neuronal Pdgfrα expression.
Conclusions: Mesenchymal deletion of Alms1 recapitulates metabolic features of AS, including fatty liver. This confirms a key role for Alms1 in the adipose lineage, where its loss is sufficient to cause systemic metabolic effects and damage to remote organs. Hyperphagia in females may depend on Alms1 deficiency in oligodendrocyte precursor cells rather than neurones. AS should be regarded as a forme fruste of lipodystrophy.
Keywords: Adipose tissue; Alms1; Alström syndrome; Diabetes; Insulin resistance; Mouse.
Copyright © 2024 The Authors. Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Robert Semple reports a relationship with AstraZeneca Pharmaceuticals LP that includes: consulting or advisory. Robert Semple reports a relationship with Amryt Pharmaceuticals Inc that includes: consulting or advisory. Robert Semple reports a relationship with Eli Lilly and Company that includes: speaking and lecture fees. Robert Semple reports a relationship with Novo Nordisk Inc that includes: speaking and lecture fees. Robert Semple reports a relationship with Novartis Pharma AG that includes: consulting or advisory. Lora Heisler reports a relationship with AstraZeneca Pharmaceuticals LP that includes: consulting or advisory. Co-author LKH is an editor for Molecular Metabolism If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Update of
-
Mesenchymal-specific Alms1 knockout in mice recapitulates key metabolic features of Alström Syndrome.bioRxiv [Preprint]. 2023 Oct 13:2023.10.12.562074. doi: 10.1101/2023.10.12.562074. bioRxiv. 2023. Update in: Mol Metab. 2024 Jun;84:101933. doi: 10.1016/j.molmet.2024.101933. PMID: 37873427 Free PMC article. Updated. Preprint.
References
-
- Paisey R.B., Steeds R., Barrett T., Williams D., Geberhiwot T., Gunay-Aygun M. GeneReviews® [Internet] University of Washington; Seattle: 2019. Alström syndrome.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

