Impaired Glycosylation of Gastric Mucins Drives Gastric Tumorigenesis and Serves as a Novel Therapeutic Target
- PMID: 38583723
- PMCID: PMC12197564
- DOI: 10.1053/j.gastro.2024.03.037
Impaired Glycosylation of Gastric Mucins Drives Gastric Tumorigenesis and Serves as a Novel Therapeutic Target
Abstract
Background & aims: Gastric cancer is often accompanied by a loss of mucin 6 (MUC6), but its pathogenic role in gastric carcinogenesis remains unclear.
Methods: Muc6 knockout (Muc6-/-) mice and Muc6-dsRED mice were newly generated. Tff1Cre, Golph3-/-, R26-Golgi-mCherry, Hes1flox/flox, Cosmcflox/flox, and A4gnt-/- mice were also used. Histology, DNA and RNA, proteins, and sugar chains were analyzed by whole-exon DNA sequence, RNA sequence, immunohistochemistry, lectin-binding assays, and liquid chromatography-mass spectrometry analysis. Gastric organoids and cell lines were used for in vitro assays and xenograft experiments.
Results: Deletion of Muc6 in mice spontaneously causes pan-gastritis and invasive gastric cancers. Muc6-deficient tumor growth was dependent on mitogen-activated protein kinase activation, mediated by Golgi stress-induced up-regulation of Golgi phosphoprotein 3. Glycomic profiling revealed aberrant expression of mannose-rich N-linked glycans in gastric tumors, detected with banana lectin in association with lack of MUC6 expression. We identified a precursor of clusterin as a binding partner of mannose glycans. Mitogen-activated protein kinase activation, Golgi stress responses, and aberrant mannose expression are found in separate Cosmc- and A4gnt-deficient mouse models that lack normal O-glycosylation. Banana lectin-drug conjugates proved an effective treatment for mannose-rich murine and human gastric cancer.
Conclusions: We propose that Golgi stress responses and aberrant glycans are important drivers of and promising new therapeutic targets for gastric cancer.
Keywords: GOLPH3; Gastric Cancer; Golgi Stress; MAPK Pathway; Muc6.
Copyright © 2024 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
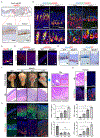



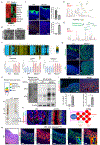
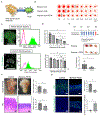

References
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–249. - PubMed
-
- Hayakawa Y, Sethi N, Sepulveda AR, et al. Oesophageal adenocarcinoma and gastric cancer: should we mind the gap? Nat Rev Cancer 2016;16:305–18. - PubMed
-
- Huang KK, Ramnarayanan K, Zhu F, et al. Genomic and Epigenomic Profiling of High-Risk Intestinal Metaplasia Reveals Molecular Determinants of Progression to Gastric Cancer. Cancer Cell 2018;33:137–150.e5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

