Utilizing Low-Dimensional Molecular Embeddings for Rapid Chemical Similarity Search
- PMID: 38585224
- PMCID: PMC10998712
- DOI: 10.1007/978-3-031-56060-6_3
Utilizing Low-Dimensional Molecular Embeddings for Rapid Chemical Similarity Search
Abstract
Nearest neighbor-based similarity searching is a common task in chemistry, with notable use cases in drug discovery. Yet, some of the most commonly used approaches for this task still leverage a brute-force approach. In practice this can be computationally costly and overly time-consuming, due in part to the sheer size of modern chemical databases. Previous computational advancements for this task have generally relied on improvements to hardware or dataset-specific tricks that lack generalizability. Approaches that leverage lower-complexity searching algorithms remain relatively underexplored. However, many of these algorithms are approximate solutions and/or struggle with typical high-dimensional chemical embeddings. Here we evaluate whether a combination of low-dimensional chemical embeddings and a k-d tree data structure can achieve fast nearest neighbor queries while maintaining performance on standard chemical similarity search benchmarks. We examine different dimensionality reductions of standard chemical embeddings as well as a learned, structurally-aware embedding-SmallSA-for this task. With this framework, searches on over one billion chemicals execute in less than a second on a single CPU core, five orders of magnitude faster than the brute-force approach. We also demonstrate that SmallSA achieves competitive performance on chemical similarity benchmarks.
Keywords: Cheminformatics; Drug discovery; Virtual screening.
Figures
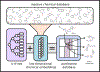

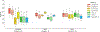

References
-
- ANNOY library. https://github.com/spotify/annoy, accessed: 2017-08-01
-
- NextMove software | Arthor, https://www.nextmovesoftware.com/arthor.html
-
- NextMove software | Arthor, https://www.nextmovesoftware.com/talks/Sayle_EvolutionVsRevolution_ICCS_...
-
- NextMove software | SmallWorld, https://www.nextmovesoftware.com/smallworld.html
-
- Achlioptas D: Database-friendly random projections: Johnson-lindenstrauss with binary coins 66(4), 671–687. 10.1016/S0022-0000(03)00025-4 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
