Repeat expansions in AR, ATXN1, ATXN2 and HTT in Norwegian patients diagnosed with amyotrophic lateral sclerosis
- PMID: 38585669
- PMCID: PMC10998343
- DOI: 10.1093/braincomms/fcae087
Repeat expansions in AR, ATXN1, ATXN2 and HTT in Norwegian patients diagnosed with amyotrophic lateral sclerosis
Abstract
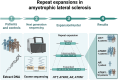
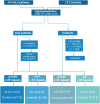
Genetic repeat expansions cause neuronal degeneration in amyotrophic lateral sclerosis as well as other neurodegenerative disorders such as spinocerebellar ataxia, Huntington's disease and Kennedy's disease. Repeat expansions in the same gene can cause multiple clinical phenotypes. We aimed to characterize repeat expansions in a Norwegian amyotrophic lateral sclerosis cohort. Norwegian amyotrophic lateral sclerosis patients (n = 414) and neurologically healthy controls adjusted for age and gender (n = 713) were investigated for repeat expansions in AR, ATXN1, ATXN2 and HTT using short read exome sequencing and the ExpansionHunter software. Five amyotrophic lateral sclerosis patients (1.2%) and two controls (0.3%) carried ≥36 repeats in HTT (P = 0.032), and seven amyotrophic lateral sclerosis patients (1.7%) and three controls (0.4%) carried ≥29 repeats in ATXN2 (P = 0.038). One male diagnosed with amyotrophic lateral sclerosis carried a pathogenic repeat expansion in AR, and his diagnosis was revised to Kennedy's disease. In ATXN1, 50 amyotrophic lateral sclerosis patients (12.1%) and 96 controls (13.5%) carried ≥33 repeats (P = 0.753). None of the patients with repeat expansions in ATXN2 or HTT had signs of Huntington's disease or spinocerebellar ataxia type 2, based on a re-evaluation of medical records. The diagnosis of amyotrophic lateral sclerosis was confirmed in all patients, with the exception of one patient who had primary lateral sclerosis. Our findings indicate that repeat expansions in HTT and ATXN2 are associated with increased likelihood of developing amyotrophic lateral sclerosis. Further studies are required to investigate the potential relationship between HTT repeat expansions and amyotrophic lateral sclerosis.
Keywords: Norway; amyotrophic lateral sclerosis; genetic risk factor; population-based study.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Goutman SA, Chen KS, Paez-Colasante X, Feldman EL. Emerging understanding of the genotype–phenotype relationship in amyotrophic lateral sclerosis. Handb Clin Neurol. 2018;148:603–623. - PubMed
-
- Mathis S, Goizet C, Soulages A, Vallat JM, Masson GL. Genetics of amyotrophic lateral sclerosis: A review. J Neurol Sci. 2019;399:217–226. - PubMed
-
- Olsen CG, Busk OL, Aanjesen TN, et al. Genetic epidemiology of amyotrophic lateral sclerosis in Norway: A 2-year population-based study. Neuroepidemiology. 2022;56(4):271–282. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
