Gut phageome: challenges in research and impact on human microbiota
- PMID: 38585689
- PMCID: PMC10995246
- DOI: 10.3389/fmicb.2024.1379382
Gut phageome: challenges in research and impact on human microbiota
Abstract
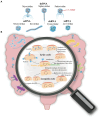
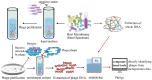
The human gut microbiome plays a critical role in maintaining our health. Fluctuations in the diversity and structure of the gut microbiota have been implicated in the pathogenesis of several metabolic and inflammatory conditions. Dietary patterns, medication, smoking, alcohol consumption, and physical activity can all influence the abundance of different types of microbiota in the gut, which in turn can affect the health of individuals. Intestinal phages are an essential component of the gut microbiome, but most studies predominantly focus on the structure and dynamics of gut bacteria while neglecting the role of phages in shaping the gut microbiome. As bacteria-killing viruses, the distribution of bacteriophages in the intestine, their role in influencing the intestinal microbiota, and their mechanisms of action remain elusive. Herein, we present an overview of the current knowledge of gut phages, their lifestyles, identification, and potential impact on the gut microbiota.
Keywords: composition; gut microbiome; gut phages; interactions; lysogenic phages; phage identification.
Copyright © 2024 Yu, Cheng, Yi, Li, Li, Liu, Liu and Kong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Role of bacteriophages in shaping gut microbial community.Gut Microbes. 2024 Jan-Dec;16(1):2390720. doi: 10.1080/19490976.2024.2390720. Epub 2024 Aug 21. Gut Microbes. 2024. PMID: 39167701 Free PMC article. Review.
-
Shining Light on Human Gut Bacteriophages.Front Cell Infect Microbiol. 2020 Sep 10;10:481. doi: 10.3389/fcimb.2020.00481. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33014897 Free PMC article. Review.
-
Gut Phageome-An Insight into the Role and Impact of Gut Microbiome and Their Correlation with Mammal Health and Diseases.Microorganisms. 2023 Sep 29;11(10):2454. doi: 10.3390/microorganisms11102454. Microorganisms. 2023. PMID: 37894111 Free PMC article. Review.
-
Microbiome-phage interactions in inflammatory bowel disease.Clin Microbiol Infect. 2023 Jun;29(6):682-688. doi: 10.1016/j.cmi.2022.08.027. Epub 2022 Oct 1. Clin Microbiol Infect. 2023. PMID: 36191844 Review.
-
Bacteriophages of the Urinary Microbiome.J Bacteriol. 2018 Mar 12;200(7):e00738-17. doi: 10.1128/JB.00738-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29378882 Free PMC article.
Cited by
-
Role of bacteriophages in shaping gut microbial community.Gut Microbes. 2024 Jan-Dec;16(1):2390720. doi: 10.1080/19490976.2024.2390720. Epub 2024 Aug 21. Gut Microbes. 2024. PMID: 39167701 Free PMC article. Review.
-
Phage treatment of multidrug-resistant bacterial infections in humans, animals, and plants: The current status and future prospects.Infect Med (Beijing). 2025 Feb 5;4(1):100168. doi: 10.1016/j.imj.2025.100168. eCollection 2025 Mar. Infect Med (Beijing). 2025. PMID: 40104270 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

