This is a preprint.
Massively parallel combination screen reveals small molecule sensitization of antibiotic-resistant Gram-negative ESKAPE pathogens
- PMID: 38585790
- PMCID: PMC10996685
- DOI: 10.1101/2024.03.26.586803
Massively parallel combination screen reveals small molecule sensitization of antibiotic-resistant Gram-negative ESKAPE pathogens
Update in
-
Large-scale combination screens reveal small-molecule sensitization of antibiotic-resistant gram-negative ESKAPE pathogens.Proc Natl Acad Sci U S A. 2025 Apr;122(13):e2402017122. doi: 10.1073/pnas.2402017122. Epub 2025 Mar 24. Proc Natl Acad Sci U S A. 2025. PMID: 40127266 Free PMC article.
Abstract
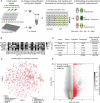
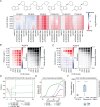
Antibiotic resistance, especially in multidrug-resistant ESKAPE pathogens, remains a worldwide problem. Combination antimicrobial therapies may be an important strategy to overcome resistance and broaden the spectrum of existing antibiotics. However, this strategy is limited by the ability to efficiently screen large combinatorial chemical spaces. Here, we deployed a high-throughput combinatorial screening platform, DropArray, to evaluate the interactions of over 30,000 compounds with up to 22 antibiotics and 6 strains of Gram-negative ESKAPE pathogens, totaling to over 1.3 million unique strain-antibiotic-compound combinations. In this dataset, compounds more frequently exhibited synergy with known antibiotics than single-agent activity. We identified a compound, P2-56, and developed a more potent analog, P2-56-3, which potentiated rifampin (RIF) activity against Acinetobacter baumannii and Klebsiella pneumoniae. Using phenotypic assays, we showed P2-56-3 disrupts the outer membrane of A. baumannii. To identify pathways involved in the mechanism of synergy between P2-56-3 and RIF, we performed genetic screens in A. baumannii. CRISPRi-induced partial depletion of lipooligosaccharide transport genes (lptA-D, lptFG) resulted in hypersensitivity to P2-56-3/RIF treatment, demonstrating the genetic dependency of P2-56-3 activity and RIF sensitization on lpt genes in A. baumannii. Consistent with outer membrane homeostasis being an important determinant of P2-56-3/RIF tolerance, knockout of maintenance of lipid asymmetry complex genes and overexpression of certain resistance-nodulation-division efflux pumps - a phenotype associated with multidrug-resistance - resulted in hypersensitivity to P2-56-3. These findings demonstrate the immense scale of phenotypic antibiotic combination screens using DropArray and the potential for such approaches to discover new small molecule synergies against multidrug-resistant ESKAPE strains.
Keywords: Acinetobacter baumannii; Biological Sciences; Gram-negative ESKAPE pathogens; Microbiology; antibiotic combinations discovery; antibiotic potentiators.
Conflict of interest statement
Competing Interests Statement: P.C.B. is a consultant to or holds equity in 10X Genomics, General Automation Lab Technologies/Isolation Bio, Celsius Therapeutics, Next Gen Diagnostics, Cache DNA, Concerto Biosciences, Stately, Ramona Optics, Bifrost Biosystems, and Amber Bio. His laboratory has received research funding from Calico Life Sciences, Merck, and Genentech for unrelated work. The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed.
Figures



References
-
- O’Neill J., Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance (2016).
-
- Tacconelli E., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). - PubMed
-
- Trusts P. C., Antibiotics Currently in Global Clinical Development (2021).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
