This is a preprint.
Temporal Single-Cell Sequencing Analysis Reveals That GPNMB-Expressing Macrophages Potentiate Muscle Regeneration
- PMID: 38585871
- PMCID: PMC10996783
- DOI: 10.21203/rs.3.rs-4108866/v1
Temporal Single-Cell Sequencing Analysis Reveals That GPNMB-Expressing Macrophages Potentiate Muscle Regeneration
Update in
-
Temporal single-cell sequencing analysis reveals that GPNMB-expressing macrophages potentiate muscle regeneration.Exp Mol Med. 2025 Jun;57(6):1232-1245. doi: 10.1038/s12276-025-01467-4. Epub 2025 Jun 9. Exp Mol Med. 2025. PMID: 40490493 Free PMC article.
Abstract
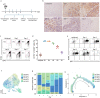
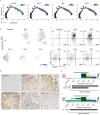
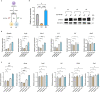
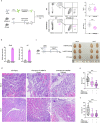
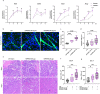
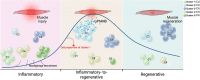
Macrophages play a crucial role in coordinating the skeletal muscle repair response, but their phenotypic diversity and the transition of specialized subsets to resolution-phase macrophages remain poorly understood. To address this issue, we induced injury and performed single-cell RNA sequencing on individual cells in skeletal muscle at different time points. Our analysis revealed a distinct macrophage subset that expressed high levels of Gpnmb and that coexpressed critical factors involved in macrophage-mediated muscle regeneration, including Igf1, Mertk, and Nr1h3. Gpnmb gene knockout inhibited macrophage-mediated efferocytosis and impaired skeletal muscle regeneration. Functional studies demonstrated that GPNMB acts directly on muscle cells in vitro and improves muscle regeneration in vivo. These findings provide a comprehensive transcriptomic atlas of macrophages during muscle injury, highlighting the key role of the GPNMB macrophage subset in regenerative processes. Targeting GPNMB signaling in macrophages could have therapeutic potential for restoring skeletal muscle integrity and homeostasis.
Conflict of interest statement
Declarations CONFLICT OF INTEREST The authors declare no competing interests.
Figures

References
-
- Giordani L. et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

