This is a preprint.
Intron-lariat spliceosomes convert lariats to true circles: implications for intron transposition
- PMID: 38585890
- PMCID: PMC10996645
- DOI: 10.1101/2024.03.26.586863
Intron-lariat spliceosomes convert lariats to true circles: implications for intron transposition
Update in
-
Intron lariat spliceosomes convert lariats to true circles: implications for intron transposition.Genes Dev. 2024 May 21;38(7-8):322-335. doi: 10.1101/gad.351764.124. Genes Dev. 2024. PMID: 38724209 Free PMC article.
Abstract
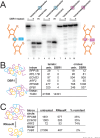
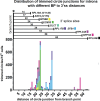
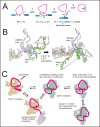
Rare, full length circular intron RNAs distinct from lariats have been reported in several species, but their biogenesis is not understood. We envision and test a hypothesis for their formation using Saccharomyces cerevisiae, documenting full length and novel processed circular RNAs from multiple introns. Evidence implicates a previously undescribed catalytic activity of the intron-lariat spliceosome (ILS) in which the 3'-OH of the lariat tail (with optional trimming and adenylation by the nuclear 3' processing machinery) attacks the branch, joining the intron 3' end to the 5' splice site in a 3'-5' linked circle. Human U2 and U12 spliceosomes produce analogous full length and processed circles. Post-splicing catalytic activity of the spliceosome may promote intron transposition during eukaryotic genome evolution.
Figures




Similar articles
-
Intron lariat spliceosomes convert lariats to true circles: implications for intron transposition.Genes Dev. 2024 May 21;38(7-8):322-335. doi: 10.1101/gad.351764.124. Genes Dev. 2024. PMID: 38724209 Free PMC article.
-
Group II intron lariat: Structural insights into the spliceosome.RNA Biol. 2015;12(9):913-7. doi: 10.1080/15476286.2015.1066956. RNA Biol. 2015. PMID: 26121424 Free PMC article. Review.
-
The circle to lariat ratio of the Ll.LtrB group II intron from Lactococcus lactis is greatly influenced by a variety of biological determinants in vivo.PLoS One. 2020 Aug 18;15(8):e0237367. doi: 10.1371/journal.pone.0237367. eCollection 2020. PLoS One. 2020. PMID: 32810148 Free PMC article.
-
Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae.Cell. 2017 Sep 21;171(1):120-132.e12. doi: 10.1016/j.cell.2017.08.029. Epub 2017 Sep 14. Cell. 2017. PMID: 28919079
-
Roles of minor spliceosome in intron recognition and the convergence with the better understood major spliceosome.Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1761. doi: 10.1002/wrna.1761. Epub 2022 Sep 2. Wiley Interdiscip Rev RNA. 2023. PMID: 36056453 Review.
References
-
- Ares M. 2012. Isolation of total RNA from yeast cell cultures. Cold Spring Harb Protoc 2012: 1082–1086. - PubMed
-
- Bertram K, Agafonov DE, Liu W-T, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. - PubMed
-
- Bertram K, El Ayoubi L, Dybkov O, Agafonov DE, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2020. Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome. Mol Cell 80: 127–139.e6. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
