This is a preprint.
Large-scale network analysis of the cerebrospinal fluid proteome identifies molecular signatures of frontotemporal lobar degeneration
- PMID: 38585969
- PMCID: PMC10996789
- DOI: 10.21203/rs.3.rs-4103685/v1
Large-scale network analysis of the cerebrospinal fluid proteome identifies molecular signatures of frontotemporal lobar degeneration
Update in
-
Large-scale network analysis of the cerebrospinal fluid proteome identifies molecular signatures of frontotemporal lobar degeneration.Nat Aging. 2025 Jun;5(6):1143-1158. doi: 10.1038/s43587-025-00878-2. Epub 2025 May 16. Nat Aging. 2025. PMID: 40380000 Free PMC article.
Abstract
The pathophysiological mechanisms driving disease progression of frontotemporal lobar degeneration (FTLD) and corresponding biomarkers are not fully understood. We leveraged aptamer-based proteomics (> 4,000 proteins) to identify dysregulated communities of co-expressed cerebrospinal fluid proteins in 116 adults carrying autosomal dominant FTLD mutations (C9orf72, GRN, MAPT) compared to 39 noncarrier controls. Network analysis identified 31 protein co-expression modules. Proteomic signatures of genetic FTLD clinical severity included increased abundance of RNA splicing (particularly in C9orf72 and GRN) and extracellular matrix (particularly in MAPT) modules, as well as decreased abundance of synaptic/neuronal and autophagy modules. The generalizability of genetic FTLD proteomic signatures was tested and confirmed in independent cohorts of 1) sporadic progressive supranuclear palsy-Richardson syndrome and 2) frontotemporal dementia spectrum syndromes. Network-based proteomics hold promise for identifying replicable molecular pathways in adults living with FTLD. 'Hub' proteins driving co-expression of affected modules warrant further attention as candidate biomarkers and therapeutic targets.
Conflict of interest statement
Competing Interests A.M.S. received research support from the NIA/NIH, the Bluefield Project to Cure FTD, and the Larry L. Hillblom Foundation. He has provided consultation to Alector, Lilly, Passage Bio, and Takeda. L.F. receives research support from the NIH. OH has received research support (for the institution) from AVID Radiopharmaceuticals, Biogen, C2N Diagnostics, Eli Lilly, Eisai, Fujirebio, GE Healthcare, and Roche. In the past 2 years, he has received consultancy/speaker fees from AC Immune, Alzpath, BioArctic, Biogen, Bristol Meyer Squibb, Cerveau, Eisai, Eli Lilly, Fujirebio, Merck, Novartis, Novo Nordisk, Roche, Sanofi and Siemens. J.S.Y. receives research support from the NIH. P.L. is a site primary investigator for clinical trials by Alector, AbbVie and Woolsey. He serves as an advisor for Retrotrope. He receives research and salary support from the NIH-NIA and the Alzheimer’s Association-Part the Cloud partnership. E.B. receives research support from the NIH and Lewy Body Dementia Association. B.C.D. is a consultant for Acadia, Alector, Arkuda, Biogen, Denali, Eisai, Genentech, Lilly, Merck, Novartis, Takeda and Wave Lifesciences; receives royalties from Cambridge University Press, Elsevier and Oxford University Press; and receives grant funding from the NIA, the National Institute of Neurological Disorders and Stroke, the National Institute of Mental Health and the Bluefield Foundation. K.D.-R. receives research support from the NIH and serves as an investigator for a clinical trial sponsored by Lawson Health Research Institute. K.F. receives research support from the NIH. J.A.F. receives research support from the NIH. N.G. has participated or is currently participating in clinical trials of anti-dementia drugs sponsored by Bristol Myers Squibb, Eli Lilly/Avid Radiopharmaceuticals, Janssen Immunotherapy, Novartis, Pfizer, Wyeth, SNIFF (The Study of Nasal Insulin to Fight Forgetfulness) and the A4 (The Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease) trial; and receives research support from Tau Consortium, the Association for Frontotemporal Dementia and the NIH. N.G.-R. receives royalties from UpToDate and has participated in multicenter therapy studies by sponsored by Biogen, TauRx, and Lilly; and receives research support from the NIH. C.M.H. is a Site PI or SubI for several industry (Alector, Janssen, Biogen, Cogito Tx) sponsored clinical trials with funding through Emory Office of Sponsored Programs. L.S.H. receives research funding from Abbvie, Acumen, Alector, Biogen, BMS, Eisai, Genentech/Roche, Janssen/J&J, Transposon, UCB, Vaccinex; and receives consulting fees from Biogen, Cortexyme, Eisai, Medscape, Prevail/Lilly. G.-Y.H. has served as an investigator for clinical trials sponsored by AstraZeneca, Eli Lilly and Roche/Genentech; and he receives research support from the Canadian Institutes of Health Research and the Alzheimer Society of British Columbia. E.D.H. receives research support from the NIH. J. Kornak has provided expert witness testimony for Teva Pharmaceuticals in Forest Laboratories Inc. et al. v. Teva Pharmaceuticals USA, Inc., case numbers 1:14-cv-00121 and 1:14-cv-00686 (D. Del. filed 31 January 2014 and 30 May 2014 regarding the drug Memantine) and for Apotex/HEC/Ezra in Novartis AG et al. v. Apotex Inc., case number 1:15-cv-975 (D. Del. filed 26 October 2015 regarding the drug Fingolimod); he has also given testimony on behalf of Puma Biotechnology in Hsingching Hsu et al, vs. Puma Biotechnology, Inc., et al. 2018 regarding the drug Neratinib; and he receives research support from the NIH. W.K. receives research funding from AstraZeneca, Biogen, Roche, the Department of Defense and the NIH. M.I.L. receives research support from the NIH. I.L.’s research is supported by the National Institutes of Health grants: 2R01AG038791–06A, U01NS100610, U01NS80818, R25NS098999; U19 AG063911–1 and 1R21NS114764–01A1; the Michael J Fox Foundation, Parkinson Foundation, Lewy Body Association, CurePSP, Roche, Abbvie, Biogen, Centogene. EIP-Pharma, Biohaven Pharmaceuticals, Novartis, Brain Neurotherapy Bio and United Biopharma SRL - UCB. She is a Scientific advisor for Amydis and Rossy Center for Progressive Supranuclear Palsy University of Toronto. She receives her salary from the University of California San Diego and as Chief Editor of Frontiers in Neurology. C.T.M receives funding from NIH and Penn Institute on Aging. M.F.M. receives research support from the NIH. A.P. receives research support from the NIH (U01 NS102035; K23 AG059891). H.P. receives research support from the NIH. L.P. receives research support from the NIH. E.M.R. receives research support from the NIH. K.R. receives research support from the NIH. E.D.R. has received research support from the NIH, the Bluefield Project to Cure Frontotemporal Dementia, the Alzheimer’s Association, the Alzheimer’s Drug Discovery Foundation, the BrightFocus Foundation, and Alector; has served as a consultant for AGTC and on a data monitoring committee for Lilly; and owns intellectual property related to tau and progranulin. R.S. receives support from the NIA, the National Institute of Neurological Disorders and Stroke, the Parkinson’s Disease Foundation and Acadia Pharmaceuticals. M.C.T. has served as an investigator for clinical trials sponsored by Biogen, Avanex, Green Valley, Roche/Genentech, Bristol Myers Squibb, Eli Lilly/Avid Radiopharmaceuticals and Janssen. She receives research support from the Canadian Institutes of Health Research. B.F.B. has served as an investigator for clinical trials sponsored by Alector, Biogen, Transposon and Cognition Therapeutics. He serves on the Scientific Advisory Board of the Tau Consortium which is funded by the Rainwater Charitable Foundation. He receives research support from NIH. H.J.R. has received research support from Biogen Pharmaceuticals, has consulting agreements with Wave Neuroscience, Ionis Pharmaceuticals, Eisai Pharmaceuticals, and Genentech, and receives research support from the NIH and the state of California. J.C.R. receives research support from the NIH and is a site principal investigator for clinical trials sponsored by Eli Lilly and Eisai. A.L.B. receives research support from the NIH, the Tau Research Consortium, the Association for Frontotemporal Degeneration, Bluefield Project to Cure Frontotemporal Dementia, Corticobasal Degeneration Solutions, the Alzheimer’s Drug Discovery Foundation and the Alzheimer’s Association. He has served as a consultant for Aeovian, AGTC, Alector, Arkuda, Arvinas, Boehringer Ingelheim, Denali, GSK, Life Edit, Humana, Oligomerix, Oscotec, Roche, TrueBinding, Wave, Merck and received research support from Biogen, Eisai and Regeneron.
Figures
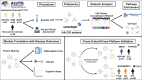



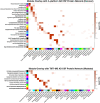
References
-
- Mackenzie IRA, Neumann M (2016) Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem 138:54–70 - PubMed
Publication types
Grants and funding
- K23 AG061253/AG/NIA NIH HHS/United States
- R01 AG058233/AG/NIA NIH HHS/United States
- R21 NS114764/NS/NINDS NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- U01 NS080818/NS/NINDS NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- R01 AG032306/AG/NIA NIH HHS/United States
- K23 AG059888/AG/NIA NIH HHS/United States
- R01 AG032289/AG/NIA NIH HHS/United States
- R01 AG062268/AG/NIA NIH HHS/United States
- U01 AG045390/AG/NIA NIH HHS/United States
- U54 NS092089/NS/NINDS NIH HHS/United States
- R25 NS098999/NS/NINDS NIH HHS/United States
- K23 AG073514/AG/NIA NIH HHS/United States
- K24 AG045333/AG/NIA NIH HHS/United States
- U01 AG079850/AG/NIA NIH HHS/United States
- U01 NS100610/NS/NINDS NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R01 AG048234/AG/NIA NIH HHS/United States
- U01 NS102035/NS/NINDS NIH HHS/United States
- R01 AG038791/AG/NIA NIH HHS/United States
- U19 AG063911/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- K23 AG059891/AG/NIA NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- R01 MH120794/MH/NIMH NIH HHS/United States
- R01 AG072475/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

