Targeting super-enhancer activity for colorectal cancer therapy
- PMID: 38586095
- PMCID: PMC10994804
- DOI: 10.62347/QKHB5897
Targeting super-enhancer activity for colorectal cancer therapy
Abstract
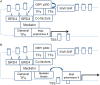
In addition to genetic variants and copy number alterations, epigenetic deregulation of oncogenes and tumor suppressors is a major contributor in cancer development and propagation. Regulatory elements for gene transcription regulation can be found in promoters which are located in the vicinity of transcription start sites but also at a distance, in enhancer sites, brought to interact with proximal sites when occupied by enhancer protein complexes. These sites provide most of the specific regulatory sequences recognized by transcription factors. A sub-set of enhancers characterized by a longer structure and stronger activity, called super-enhancers, are critical for the expression of specific genes, usually associated with individual cell type identity and function. Super-enhancers show deregulation in cancer, which may have profound repercussions for cancer cell survival and response to therapy. Dysfunction of super-enhancers may result from multiple mechanisms that include changes in their sequence, alterations in the topological neighborhoods where they belong, and alterations in the proteins that mediate their function, such as transcription factors and epigenetic modifiers. These can become potential targets for therapeutic interventions. Genes that are targets of super-enhancers are cell and cancer type specific and could also be of interest for therapeutic targeting. In colorectal cancer, a super-enhancer regulated and over-expressed oncogene is MYC, under the influence of the WNT/β-catenin pathway. Identification and targeting of additional oncogenes regulated by super-enhancers in colorectal cancer may pave the way for combination therapies targeting the super-enhancer machinery and signal transduction pathways that regulate the specific transcription factors operative on them.
Keywords: Epigenetics; enhancers; gene regulation; histone acetylation; promoters.
AJTR Copyright © 2024.
Conflict of interest statement
None.
Figures
Similar articles
-
The molecular understanding of super-enhancer dysregulation in cancer.Nagoya J Med Sci. 2022 May;84(2):216-229. doi: 10.18999/nagjms.84.2.216. Nagoya J Med Sci. 2022. PMID: 35967935 Free PMC article. Review.
-
Oncogenic super-enhancers in cancer: mechanisms and therapeutic targets.Cancer Metastasis Rev. 2023 Jun;42(2):471-480. doi: 10.1007/s10555-023-10103-4. Epub 2023 Apr 14. Cancer Metastasis Rev. 2023. PMID: 37059907 Free PMC article. Review.
-
Super-enhancers in cancer.Pharmacol Ther. 2019 Jul;199:129-138. doi: 10.1016/j.pharmthera.2019.02.014. Epub 2019 Mar 16. Pharmacol Ther. 2019. PMID: 30885876 Review.
-
Super-enhancers and novel therapeutic targets in colorectal cancer.Cell Death Dis. 2022 Mar 11;13(3):228. doi: 10.1038/s41419-022-04673-4. Cell Death Dis. 2022. PMID: 35277481 Free PMC article. Review.
-
Targeting Super-Enhancer-Associated Oncogenes in Osteosarcoma with THZ2, a Covalent CDK7 Inhibitor.Clin Cancer Res. 2020 Jun 1;26(11):2681-2692. doi: 10.1158/1078-0432.CCR-19-1418. Epub 2020 Jan 14. Clin Cancer Res. 2020. PMID: 31937612
Cited by
-
Therapeutic opportunities for hypermutated urothelial carcinomas beyond immunotherapy.Oncoscience. 2024 Apr 25;11:36-37. doi: 10.18632/oncoscience.596. eCollection 2024. Oncoscience. 2024. PMID: 38699226 Free PMC article. No abstract available.
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. - PubMed
-
- Bando H, Ohtsu A, Yoshino T. Therapeutic landscape and future direction of metastatic colorectal cancer. Nat Rev Gastroenterol Hepatol. 2023;20:306–322. - PubMed
-
- Yaeger R, Weiss J, Pelster MS, Spira AI, Barve M, Ou SI, Leal TA, Bekaii-Saab TS, Paweletz CP, Heavey GA, Christensen JG, Velastegui K, Kheoh T, Der-Torossian H, Klempner SJ. Adagrasib with or without cetuximab in colorectal cancer with mutated KRAS G12C. N Engl J Med. 2023;388:44–54. - PMC - PubMed
-
- Sakamoto Y, Bando H, Nakamura Y, Hasegawa H, Kuwaki T, Okamoto W, Taniguchi H, Aoyagi Y, Miki I, Uchigata H, Kuramoto N, Fuse N, Yoshino T, Ohtsu A. Trajectory for the regulatory approval of a combination of pertuzumab plus trastuzumab for pre-treated HER2-positive metastatic colorectal cancer using real-world data. Clin Colorectal Cancer. 2023;22:45–52. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
