Diagnosis of malignant body fluids via cancer-universal methylation in cell-free DNA
- PMID: 38587071
- PMCID: PMC11128206
- DOI: 10.1172/jci.insight.175482
Diagnosis of malignant body fluids via cancer-universal methylation in cell-free DNA
Abstract
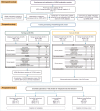
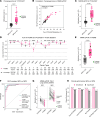
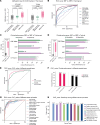
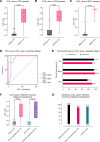
BACKGROUNDDifferentiating malignant from nonmalignant body fluids remains a clinical challenge because of the unsatisfying performance of conventional cytology. We aimed to improve the sensitivity and ubiquity of cancer cell detection by assaying universal cancer-only methylation (UCOM) markers in supernatant cell-free DNA (cfDNA).METHODSAn observational prospective cohort including 1,321 nonmalignant and malignant body fluids of multiple cancers was used to develop and validate a cfDNA UCOM methylation diagnostic assay. All samples were divided into 2 portions for cytology and supernatant cfDNA methylation analysis.RESULTSThe significant hypermethylation of a potentially novel UCOM marker, TAGMe, together with the formerly reported PCDHGB7, was identified in the cfDNA of malignant body fluid samples. The combined model, cell-free cancer-universal methylation (CUE), was developed and validated in a prospective multicancer cohort with markedly elevated sensitivity and specificity, and was further verified in a set containing additional types of malignant body fluids and metastases. In addition, it remained hypersensitive in detecting cancer cells in cytologically negative malignant samples.CONCLUSIONcfDNA methylation markers are robust in detecting tumor cells and are applicable to diverse body fluids and tumor types, providing a feasible complement to current cytology-based diagnostic analyses.TRIAL REGISTRATIONThis study was registered at Chictr.org.cn (ChiCTR2200060532).FUNDINGNational Natural Science Foundation of China (32270645, 31872814, 32000505, 82170088), the National Key R&D Program of Ningxia Hui Autonomous region (2022BEG01003), Shanghai Municipal Key Clinical Specialty (shslczdzk02201), Science and Technology Commission of Shanghai Municipality (20DZ2261200, 20DZ2254400), and Major Special Projects of Basic Research of Shanghai Science and Technology Commission (18JC1411101).
Keywords: Cancer; Epigenetics; Genetics; Molecular diagnosis; Oncology.
Figures
Similar articles
-
Targeted deep sequencing of cell-free DNA in serous body cavity fluids with malignant, suspicious, and benign cytology.Cancer Cytopathol. 2020 Jan;128(1):43-56. doi: 10.1002/cncy.22205. Epub 2019 Nov 21. Cancer Cytopathol. 2020. PMID: 31751001
-
Liquid biopsies: DNA methylation analyses in circulating cell-free DNA.J Genet Genomics. 2018 Apr 20;45(4):185-192. doi: 10.1016/j.jgg.2018.02.007. Epub 2018 Mar 8. J Genet Genomics. 2018. PMID: 29706556 Review.
-
Unintrusive multi-cancer detection by circulating cell-free DNA methylation sequencing (THUNDER): development and independent validation studies.Ann Oncol. 2023 May;34(5):486-495. doi: 10.1016/j.annonc.2023.02.010. Epub 2023 Feb 26. Ann Oncol. 2023. PMID: 36849097
-
Peritoneal cell-free DNA as a sensitive biomarker for detection of peritoneal metastasis in colorectal cancer: a prospective diagnostic study: A prospective diagnostic study.Clin Epigenetics. 2023 Apr 18;15(1):65. doi: 10.1186/s13148-023-01479-9. Clin Epigenetics. 2023. PMID: 37072801 Free PMC article. Clinical Trial.
-
Analysis of DNA Methylation Status in Bodily Fluids for Early Detection of Cancer.Int J Mol Sci. 2017 Mar 30;18(4):735. doi: 10.3390/ijms18040735. Int J Mol Sci. 2017. PMID: 28358330 Free PMC article. Review.
Cited by
-
Hypermethylated TAGMe as a universal-cancer-only methylation marker and its application in diagnosis and recurrence monitoring of urothelial carcinoma.J Transl Med. 2024 Jul 2;22(1):608. doi: 10.1186/s12967-024-05420-3. J Transl Med. 2024. PMID: 38956589 Free PMC article.
-
Deregulated methylation and expression of PCDHGB7 in patients with non-small cell lung cancer: a novel prognostic and immunological biomarker.Front Immunol. 2025 Jan 30;16:1516628. doi: 10.3389/fimmu.2025.1516628. eCollection 2025. Front Immunol. 2025. PMID: 39949775 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

