A complement C4-derived glycopeptide is a biomarker for PMM2-CDG
- PMID: 38587076
- PMCID: PMC7615924
- DOI: 10.1172/jci.insight.172509
A complement C4-derived glycopeptide is a biomarker for PMM2-CDG
Abstract
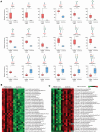
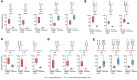
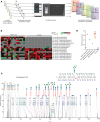
BACKGROUNDDiagnosis of PMM2-CDG, the most common congenital disorder of glycosylation (CDG), relies on measuring carbohydrate-deficient transferrin (CDT) and genetic testing. CDT tests have false negatives and may normalize with age. Site-specific changes in protein N-glycosylation have not been reported in sera in PMM2-CDG.METHODSUsing multistep mass spectrometry-based N-glycoproteomics, we analyzed sera from 72 individuals to discover and validate glycopeptide alterations. We performed comprehensive tandem mass tag-based discovery experiments in well-characterized patients and controls. Next, we developed a method for rapid profiling of additional samples. Finally, targeted mass spectrometry was used for validation in an independent set of samples in a blinded fashion.RESULTSOf the 3,342 N-glycopeptides identified, patients exhibited decrease in complex-type N-glycans and increase in truncated, mannose-rich, and hybrid species. We identified a glycopeptide from complement C4 carrying the glycan Man5GlcNAc2, which was not detected in controls, in 5 patients with normal CDT results, including 1 after liver transplant and 2 with a known genetic variant associated with mild disease, indicating greater sensitivity than CDT. It was detected by targeted analysis in 2 individuals with variants of uncertain significance in PMM2.CONCLUSIONComplement C4-derived Man5GlcNAc2 glycopeptide could be a biomarker for accurate diagnosis and therapeutic monitoring of patients with PMM2-CDG and other CDGs.FUNDINGU54NS115198 (Frontiers in Congenital Disorders of Glycosylation: NINDS; NCATS; Eunice Kennedy Shriver NICHD; Rare Disorders Consortium Disease Network); K08NS118119 (NINDS); Minnesota Partnership for Biotechnology and Medical Genomics; Rocket Fund; R01DK099551 (NIDDK); Mayo Clinic DERIVE Office; Mayo Clinic Center for Biomedical Discovery; IA/CRC/20/1/600002 (Center for Rare Disease Diagnosis, Research and Training; DBT/Wellcome Trust India Alliance).
Keywords: Genetic diseases; Genetics; Glycobiology; Metabolism; Proteomics.
Figures



References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

