ARID1A regulates DNA repair through chromatin organization and its deficiency triggers DNA damage-mediated anti-tumor immune response
- PMID: 38587186
- PMCID: PMC11162808
- DOI: 10.1093/nar/gkae233
ARID1A regulates DNA repair through chromatin organization and its deficiency triggers DNA damage-mediated anti-tumor immune response
Abstract
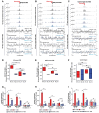
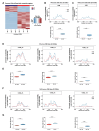
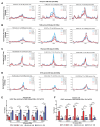
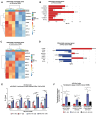
AT-rich interaction domain protein 1A (ARID1A), a SWI/SNF chromatin remodeling complex subunit, is frequently mutated across various cancer entities. Loss of ARID1A leads to DNA repair defects. Here, we show that ARID1A plays epigenetic roles to promote both DNA double-strand breaks (DSBs) repair pathways, non-homologous end-joining (NHEJ) and homologous recombination (HR). ARID1A is accumulated at DSBs after DNA damage and regulates chromatin loops formation by recruiting RAD21 and CTCF to DSBs. Simultaneously, ARID1A facilitates transcription silencing at DSBs in transcriptionally active chromatin by recruiting HDAC1 and RSF1 to control the distribution of activating histone marks, chromatin accessibility, and eviction of RNAPII. ARID1A depletion resulted in enhanced accumulation of micronuclei, activation of cGAS-STING pathway, and an increased expression of immunomodulatory cytokines upon ionizing radiation. Furthermore, low ARID1A expression in cancer patients receiving radiotherapy was associated with higher infiltration of several immune cells. The high mutation rate of ARID1A in various cancer types highlights its clinical relevance as a promising biomarker that correlates with the level of immune regulatory cytokines and estimates the levels of tumor-infiltrating immune cells, which can predict the response to the combination of radio- and immunotherapy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. - PubMed
-
- Lieber M.R., Wilson T.E. SnapShot: nonhomologous DNA end joining (NHEJ). Cell. 2010; 142:496–496. - PubMed
-
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

