Novel WRN Helicase Inhibitors Selectively Target Microsatellite-Unstable Cancer Cells
- PMID: 38587317
- PMCID: PMC7616858
- DOI: 10.1158/2159-8290.CD-24-0052
Novel WRN Helicase Inhibitors Selectively Target Microsatellite-Unstable Cancer Cells
Abstract
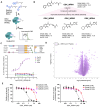
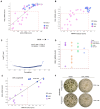
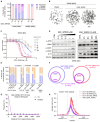
Microsatellite-unstable (MSI) cancers require WRN helicase to resolve replication stress due to expanded DNA (TA)n dinucleotide repeats. WRN is a promising synthetic lethal target for MSI tumors, and WRN inhibitors are in development. In this study, we used CRISPR-Cas9 base editing to map WRN residues critical for MSI cells, validating the helicase domain as the primary drug target. Fragment-based screening led to the development of potent and highly selective WRN helicase covalent inhibitors. These compounds selectively suppressed MSI model growth in vitro and in vivo by mimicking WRN loss, inducing DNA double-strand breaks at expanded TA repeats and DNA damage. Assessment of biomarkers in preclinical models linked TA-repeat expansions and mismatch repair alterations to compound activity. Efficacy was confirmed in immunotherapy-resistant organoids and patient-derived xenograft models. The discovery of potent, selective covalent WRN inhibitors provides proof of concept for synthetic lethal targeting of WRN in MSI cancer and tools to dissect WRN biology. Significance: We report the discovery and characterization of potent, selective WRN helicase inhibitors for MSI cancer treatment, with biomarker analysis and evaluation of efficacy in vivo and in immunotherapy-refractory preclinical models. These findings pave the way to translate WRN inhibition into MSI cancer therapies and provide tools to investigate WRN biology. See related commentary by Wainberg, p. 1369.
©2024 American Association for Cancer Research.
Conflict of interest statement
AstraZeneca, GlaxoSmithKline, and Astex Pharmaceuticals have awarded MJG research grants. Additionally, MJG and EV are founders and advisors at Mosaic Therapeutics. GP serves in a consultant role for Mosaic Therapeutics. All authors listed with an affiliation to GSK are GlaxoSmithKline (GSK) employees except JEC and YP, who are no longer GSK employees. SV is currently employed at Astrazeneca.
Figures



References
-
- Toh J, Spring K. Ann Oncol. Vol. 30. Elsevier BV; 2019. Microsatellite Instability (MSI) status and prognosis in colorectal cancer: Meta-analysis; v220.
-
- Cohen R, Rousseau B, Vidal J, Colle R, Diaz LA, Jr, André T. Immune Checkpoint Inhibition in Colorectal Cancer: Microsatellite Instability and Beyond. Target Oncol. 2020;15:11–24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

