Recombinational exchange of M-fibril and T-pilus genes generates extensive cell surface diversity in the global group A Streptococcus population
- PMID: 38587426
- PMCID: PMC11078000
- DOI: 10.1128/mbio.00693-24
Recombinational exchange of M-fibril and T-pilus genes generates extensive cell surface diversity in the global group A Streptococcus population
Abstract
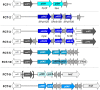
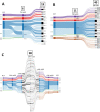
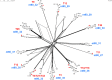
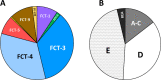
Among genes present in all group A streptococci (GAS), those encoding M-fibril and T-pilus proteins display the highest levels of sequence diversity, giving rise to the two primary serological typing schemes historically used to define strain. A new genotyping scheme for the pilin adhesin and backbone genes is developed and, when combined with emm typing, provides an account of the global GAS strain population. Cluster analysis based on nucleotide sequence similarity assigns most T-serotypes to discrete pilin backbone sequence clusters, yet the established T-types correspond to only half the clusters. The major pilin adhesin and backbone sequence clusters yield 98 unique combinations, defined as "pilin types." Numerous horizontal transfer events that involve pilin or emm genes generate extensive antigenic and functional diversity on the bacterial cell surface and lead to the emergence of new strains. Inferred pilin genotypes applied to a meta-analysis of global population-based collections of pharyngitis and impetigo isolates reveal highly significant associations between pilin genotypes and GAS infection at distinct ecological niches, consistent with a role for pilin gene products in adaptive evolution. Integration of emm and pilin typing into open-access online tools (pubmlst.org) ensures broad utility for end-users wanting to determine the architecture of M-fibril and T-pilus genes from genome assemblies.IMPORTANCEPrecision in defining the variant forms of infectious agents is critical to understanding their population biology and the epidemiology of associated diseases. Group A Streptococcus (GAS) is a global pathogen that causes a wide range of diseases and displays a highly diverse cell surface due to the antigenic heterogeneity of M-fibril and T-pilus proteins which also act as virulence factors of varied functions. emm genotyping is well-established and highly utilized, but there is no counterpart for pilin genes. A global GAS collection provides the basis for a comprehensive pilin typing scheme, and online tools for determining emm and pilin genotypes are developed. Application of these tools reveals the expansion of structural-functional diversity among GAS via horizontal gene transfer, as evidenced by unique combinations of surface protein genes. Pilin and emm genotype correlations with superficial throat vs skin infection provide new insights on the molecular determinants underlying key ecological and epidemiological trends.
Keywords: cell surface proteins; genotyping; group A streptococcus; molecular epidemiology; pili; population biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bryant AE, Stevens DL. 2020. Streptococcus pyogenes. In Mandell GL, Douglas RG, Dolin R (ed), Principles and practice of infectious diseases, 9th ed. Churchill Livingstone, Philadelphia.
-
- Johnson DR, Kaplan EL, VanGheem A, Facklam RR, Beall B. 2006. Characterization of group A Streptococci (Streptococcus pyogenes): correlation of M-protein and emm-gene type with T-protein agglutination pattern and serum opacity factor. J Med Microbiol 55:157–164. doi:10.1099/jmm.0.46224-0 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

