Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer
- PMID: 38588697
- PMCID: PMC11111406
- DOI: 10.1038/s41586-024-07379-z
Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer
Erratum in
-
Author Correction: Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer.Nature. 2024 Nov;635(8040):E12. doi: 10.1038/s41586-024-08084-7. Nature. 2024. PMID: 39533066 Free PMC article. No abstract available.
Abstract
Broad-spectrum RAS inhibition has the potential to benefit roughly a quarter of human patients with cancer whose tumours are driven by RAS mutations1,2. RMC-7977 is a highly selective inhibitor of the active GTP-bound forms of KRAS, HRAS and NRAS, with affinity for both mutant and wild-type variants3. More than 90% of cases of human pancreatic ductal adenocarcinoma (PDAC) are driven by activating mutations in KRAS4. Here we assessed the therapeutic potential of RMC-7977 in a comprehensive range of PDAC models. We observed broad and pronounced anti-tumour activity across models following direct RAS inhibition at exposures that were well-tolerated in vivo. Pharmacological analyses revealed divergent responses to RMC-7977 in tumour versus normal tissues. Treated tumours exhibited waves of apoptosis along with sustained proliferative arrest, whereas normal tissues underwent only transient decreases in proliferation, with no evidence of apoptosis. In the autochthonous KPC mouse model, RMC-7977 treatment resulted in a profound extension of survival followed by on-treatment relapse. Analysis of relapsed tumours identified Myc copy number gain as a prevalent candidate resistance mechanism, which could be overcome by combinatorial TEAD inhibition in vitro. Together, these data establish a strong preclinical rationale for the use of broad-spectrum RAS-GTP inhibition in the setting of PDAC and identify a promising candidate combination therapeutic regimen to overcome monotherapy resistance.
© 2024. The Author(s).
Conflict of interest statement
J.J., Y.W., B.L., M.M., S.C., L.J., X.W., Y.C.Y., C. Helland, H.C., Y.G., K.M., R.Z., J.B., E.Q., Z.W., J.A.M.S., M.H., D.W. and M. Singh are employees and stockholders of Revolution Medicines. S.W.L., R.H.V., B.Z.S., A.J.A., C.J.D. and K.P.O. received research funding from Revolution Medicines. A.C. is founder, equity holder and consultant of DarwinHealth, a company that has licensed some of the algorithms used in this manuscript from Columbia University. Columbia University is also an equity holder in DarwinHealth. L.T. is an employee of DarwinHealth. J.M.C. receives research funding to his institution from Merus, Roche and Bristol Myers Squibb. He receives research support from Merck, AstraZeneca, Esperas Pharma, Bayer, Tesaro, Arcus Biosciences and Apexigen; he has also received honoraria for being on the advisory boards of Syros Pharmaceuticals, Incyte and Blueprint Medicines. B.M.W. has received research funding from Celgene, Eli Lilly, Novartis and Revolution Medicine and has consulted for Celgene, GRAIL and Mirati. A.J.A. has consulted for Anji Pharmaceuticals, Affini-T Therapeutics, Arrakis Therapeutics, AstraZeneca, Boehringer Ingelheim, Oncorus, Merck & Co., Mirati Therapeutics, Nimbus Therapeutics, Plexium, Revolution Medicines, Reactive Biosciences, Riva Therapeutics, Servier Pharmaceuticals, Syros Pharmaceuticals, T-knife Therapeutics, Third Rock Ventures and Ventus Therapeutics. A.J.A. holds equity in Riva Therapeutics. A.J.A. has research funding from Bristol Myers Squibb, Deerfield, Eli Lilly, Mirati Therapeutics, Novartis, Novo Ventures, Revolution Medicines and Syros Pharmaceuticals. While not directly related to this manuscript, S.W.L. declares competing interests outside consultancy and equity for Oric Pharmaceuticals, Blueprint Medicines, Mirimus, Senecea Therapeutics, Faeth Therapeutics and PMV Pharmaceuticals, and outside consultancy (no equity) for Fate Therapeutics. C.J.D. is a consultant/advisory board member for Cullgen, Deciphera Pharmaceuticals, Eli Lilly, Mirati Therapeutics, Reactive Biosciences, Revolution Medicines, Ribometrics, Sanofi and SHY Therapeutics. C.J.D. has received research funding support from Deciphera Pharmaceuticals, Mirati Therapeutics, Reactive Biosciences, and SpringWorks Therapeutics. R.H.V. has received consulting fees from BMS, is an inventor on patents relating to cancer cellular immunotherapy, cancer vaccines and KRAS immune epitopes, and receives royalties from Children’s Hospital Boston for a licensed research-only monoclonal antibody. U.N.W., C.J.D. and K.P.O. were named as inventors on a patent application filed by Columbia University pertaining to the treatment of RAS inhibitor resistant tumours. The other authors declare no competing interests.
Figures

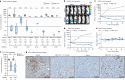
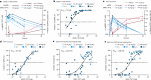
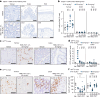





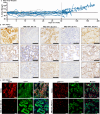

References
MeSH terms
Substances
Grants and funding
- U01 CA272610/CA/NCI NIH HHS/United States
- P50 CA257911/CA/NCI NIH HHS/United States
- R01 CA266558/CA/NCI NIH HHS/United States
- T32 CA009156/CA/NCI NIH HHS/United States
- R35 CA232113/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- T32 CA009140/CA/NCI NIH HHS/United States
- S10 OD012351/OD/NIH HHS/United States
- T32 GM119999/GM/NIGMS NIH HHS/United States
- R01 CA229803/CA/NCI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- U01 CA217858/CA/NCI NIH HHS/United States
- S10 OD021764/OD/NIH HHS/United States
- P01 CA129243/CA/NCI NIH HHS/United States
- R01 CA215607/CA/NCI NIH HHS/United States
- F31 CA275260/CA/NCI NIH HHS/United States
- U01 CA274312/CA/NCI NIH HHS/United States
- F32 CA232529/CA/NCI NIH HHS/United States
- S10 OD032433/OD/NIH HHS/United States
- P30 DK132710/DK/NIDDK NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 CA276268/CA/NCI NIH HHS/United States
- T32 CA071341/CA/NCI NIH HHS/United States
- K08 AI106953/AI/NIAID NIH HHS/United States
- U54 CA274506/CA/NCI NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

