Accelerated DNA replication fork speed due to loss of R-loops in myelodysplastic syndromes with SF3B1 mutation
- PMID: 38589367
- PMCID: PMC11001894
- DOI: 10.1038/s41467-024-46547-7
Accelerated DNA replication fork speed due to loss of R-loops in myelodysplastic syndromes with SF3B1 mutation
Abstract
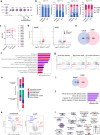
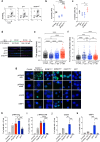
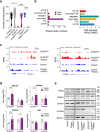
Myelodysplastic syndromes (MDS) with mutated SF3B1 gene present features including a favourable outcome distinct from MDS with mutations in other splicing factor genes SRSF2 or U2AF1. Molecular bases of these divergences are poorly understood. Here we find that SF3B1-mutated MDS show reduced R-loop formation predominating in gene bodies associated with intron retention reduction, not found in U2AF1- or SRSF2-mutated MDS. Compared to erythroblasts from SRSF2- or U2AF1-mutated patients, SF3B1-mutated erythroblasts exhibit augmented DNA synthesis, accelerated replication forks, and single-stranded DNA exposure upon differentiation. Importantly, histone deacetylase inhibition using vorinostat restores R-loop formation, slows down DNA replication forks and improves SF3B1-mutated erythroblast differentiation. In conclusion, loss of R-loops with associated DNA replication stress represents a hallmark of SF3B1-mutated MDS ineffective erythropoiesis, which could be used as a therapeutic target.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

