Yeast TLDc domain proteins regulate assembly state and subcellular localization of the V-ATPase
- PMID: 38589611
- PMCID: PMC11066047
- DOI: 10.1038/s44318-024-00097-2
Yeast TLDc domain proteins regulate assembly state and subcellular localization of the V-ATPase
Abstract
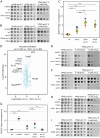
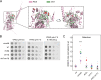
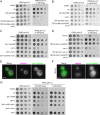
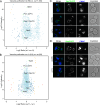
Yeast vacuoles perform crucial cellular functions as acidic degradative organelles, storage compartments, and signaling hubs. These functions are mediated by important protein complexes, including the vacuolar-type H+-ATPase (V-ATPase), responsible for organelle acidification. To gain a more detailed understanding of vacuole function, we performed cross-linking mass spectrometry on isolated vacuoles, detecting many known as well as novel protein-protein interactions. Among these, we identified the uncharacterized TLDc-domain-containing protein Rtc5 as a novel interactor of the V-ATPase. We further analyzed the influence of Rtc5 and of Oxr1, the only other yeast TLDc-domain-containing protein, on V-ATPase function. We find that both Rtc5 and Oxr1 promote the disassembly of the vacuolar V-ATPase in vivo, counteracting the role of the RAVE complex, a V-ATPase assembly chaperone. Furthermore, Oxr1 is necessary for the retention of a Golgi-specific subunit of the V-ATPase in this compartment. Collectively, our results shed light on the in vivo roles of yeast TLDc-domain proteins as regulators of the V-ATPase, highlighting the multifaceted regulation of this crucial protein complex.
Keywords: Cross-linking Mass Spectrometry; TLDc; V-ATPase; Vacuole.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Interaction between the yeast RAVE complex and Vph1-containing Vo sectors is a central glucose-sensitive interaction required for V-ATPase reassembly.J Biol Chem. 2020 Feb 21;295(8):2259-2269. doi: 10.1074/jbc.RA119.011522. Epub 2020 Jan 15. J Biol Chem. 2020. PMID: 31941791 Free PMC article.
-
Yeast phosphofructokinase-1 subunit Pfk2p is necessary for pH homeostasis and glucose-dependent vacuolar ATPase reassembly.J Biol Chem. 2014 Jul 11;289(28):19448-57. doi: 10.1074/jbc.M114.569855. Epub 2014 May 23. J Biol Chem. 2014. PMID: 24860096 Free PMC article.
-
Defining steps in RAVE-catalyzed V-ATPase assembly using purified RAVE and V-ATPase subcomplexes.J Biol Chem. 2021 Jan-Jun;296:100703. doi: 10.1016/j.jbc.2021.100703. Epub 2021 Apr 22. J Biol Chem. 2021. PMID: 33895134 Free PMC article.
-
The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase.Microbiol Mol Biol Rev. 2006 Mar;70(1):177-91. doi: 10.1128/MMBR.70.1.177-191.2006. Microbiol Mol Biol Rev. 2006. PMID: 16524922 Free PMC article. Review.
-
V-ATPase, ScNhx1p and yeast vacuole fusion.J Genet Genomics. 2012 Apr 20;39(4):167-71. doi: 10.1016/j.jgg.2012.02.001. Epub 2012 Feb 10. J Genet Genomics. 2012. PMID: 22546538 Review.
Cited by
-
Knockout of the V-ATPase interacting protein Tldc2 in B-type kidney intercalated cells impairs urine alkalinization.Am J Physiol Renal Physiol. 2025 Jun 1;328(6):F890-F906. doi: 10.1152/ajprenal.00363.2024. Epub 2025 May 13. Am J Physiol Renal Physiol. 2025. PMID: 40358928 Free PMC article.
-
V-ATPase Disassembly at the Yeast Lysosome-Like Vacuole Is a Phenotypic Driver of Lysosome Dysfunction in Replicative Aging.Aging Cell. 2025 May;24(5):e14487. doi: 10.1111/acel.14487. Epub 2025 Jan 16. Aging Cell. 2025. PMID: 39817304 Free PMC article.
-
Interaction of yeast V-ATPase with TLDc protein Rtc5p.bioRxiv [Preprint]. 2025 May 24:2025.05.24.655954. doi: 10.1101/2025.05.24.655954. bioRxiv. 2025. PMID: 40475620 Free PMC article. Preprint.
-
V-ATPase Disassembly at the Yeast Lysosome-Like Vacuole Is a Phenotypic Driver of Lysosome Dysfunction in Replicative Aging.bioRxiv [Preprint]. 2024 Dec 13:2024.07.23.604825. doi: 10.1101/2024.07.23.604825. bioRxiv. 2024. Update in: Aging Cell. 2025 May;24(5):e14487. doi: 10.1111/acel.14487. PMID: 39091794 Free PMC article. Updated. Preprint.
-
TBC1D24 interacts with the v-ATPase and regulates intraorganellar pH in neurons.iScience. 2024 Dec 1;28(1):111515. doi: 10.1016/j.isci.2024.111515. eCollection 2025 Jan 17. iScience. 2024. PMID: 39758816 Free PMC article.
References
-
- Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, et al. A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics. 2008;180:2251–2266. doi: 10.1534/genetics.108.092577. - DOI - PMC - PubMed
-
- Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35:563–573 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

