Tubulin alpha-1b chain was identified as a prognosis and immune biomarker in pan-cancer combing with experimental validation in breast cancer
- PMID: 38589634
- PMCID: PMC11001892
- DOI: 10.1038/s41598-024-58982-z
Tubulin alpha-1b chain was identified as a prognosis and immune biomarker in pan-cancer combing with experimental validation in breast cancer
Abstract
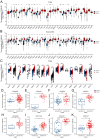
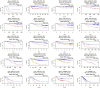
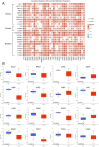
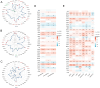
The α-tubulin subtype, Tubulin α-1b chain (TUBA1B), has been shown to influence immune cell infiltration, cancer growth, and survival across various malignancies. However, a comprehensive study has not yet been undertaken examining the immunological and predictive effects of TUBA1B in a pan-carcinoma context. Using data from TCGA, GEO, and other databases, we analyzed TUBA1B expression across various carcinoma types using transcriptional profiling, prognostic implications, genetic and epigenetic alterations, methylation patterns, and immunological significance. To validate our findings, we conducted Western blot analysis to assess TUBA1B protein levels in matched breast cancer tissue samples and performed CCK-8 proliferation assay, flow cytometry, transwell invasion, and migration assays to comprehensively examine the functional impact of TUBA1B on breast cancer cells. Our pan-cancer analysis found TUBA1B upregulation across most tumor types, with varying expression patterns in distinct immune and molecular subtypes. High TUBA1B expression was an independent risk factor and associated with poor prognoses in several cancers, including BRCA, KICH, LGG, LUAD, and MESO. TUBA1B also demonstrates moderate to high diagnostic accuracy in most tumor types. Increased m6A methylation levels were observed in the TUBA1B gene, while its promoter region displayed low methylation levels. TUBA1B's expression impacted some cancers by elevating tumor mutation burden, microsatellite instability, neoantigen formation, immune cell infiltration, and the modulation of immune checkpoints. Functional enrichment analysis highlights TUBA1B's involvement in important cellular processes such as the cell cycle, p53 signaling, cell senescence, programmed cell death, and the regulation of immune-related pathways. Moreover, our study reveals higher TUBA1B protein expression in breast cancer tissues compared to adjacent tissues. In vitro experiments confirm that TUBA1B deletion reduces breast cancer cell proliferation, invasion, and migration while increasing apoptosis. In conclusion, our study suggests that TUBA1B could potentially serve as a diagnostic marker for predicting cancer immunological profiles and survival outcomes and shed light on the expression and role of TUBA1B in breast cancer, providing a solid foundation for considering it as a promising therapeutic target for breast cancer patient treatment.
Keywords: Methylation; Pan-cancer; Prognosis; TUBA1B; Tumor immunity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- XJ2023G169/Xinjiang Uygur Autonomous Region Graduate Innovation Program
- 2022TSYCCX0029/Xinjiang Uygur Autonomous Region Youth Science and Technology Top-notch Talent Program
- 32260186/National Natural Science Foundation of China
- 2022E02136/Regional Collaborative Innovation Special Project (Science and Technology Assistance to Xinjiang Program)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

