A missense mutation in human INSC causes peripheral neuropathy
- PMID: 38589651
- PMCID: PMC11099080
- DOI: 10.1038/s44321-024-00062-w
A missense mutation in human INSC causes peripheral neuropathy
Abstract
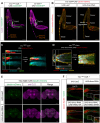
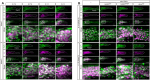
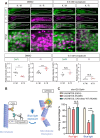
PAR3/INSC/LGN form an evolutionarily conserved complex required for asymmetric cell division in the developing brain, but its post-developmental function and disease relevance in the peripheral nervous system (PNS) remains unknown. We mapped a new locus for axonal Charcot-Marie-Tooth disease (CMT2) and identified a missense mutation c.209 T > G (p.Met70Arg) in the INSC gene. Modeling the INSCM70R variant in Drosophila, we showed that it caused proprioceptive defects in adult flies, leading to gait defects resembling those in CMT2 patients. Cellularly, PAR3/INSC/LGN dysfunction caused tubulin aggregation and necrotic neurodegeneration, with microtubule-stabilizing agents rescuing both morphological and functional defects of the INSCM70R mutation in the PNS. Our findings underscore the critical role of the PAR3/INSC/LGN machinery in the adult PNS and highlight a potential therapeutic target for INSC-associated CMT2.
Keywords: Inscuteable; Charcot–Marie–Tooth Neuropathy Type 2; Microtubule-Stabilizing Agents; Necrosis; Proprioception.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Ackerley S, James PA, Kalli A, French S, Davies KE, Talbot K. A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Hum Mol Genet. 2006;15:347–354. doi: 10.1093/hmg/ddi452. - DOI - PubMed
-
- Banerjee A, Wu S, Cheng L, Aw SS (2020) Fully automated leg tracking in freely moving insects using feature learning leg segmentation and tracking (FLLIT). J Vis Exp 158:e61012 - PubMed
-
- Blair SS. Dissection of imaginal discs in Drosophila. CSH Protoc. 2007;2007:pdb.prot4794. - PubMed
MeSH terms
Substances
Grants and funding
- 111-2320-B-002-049-MY3/National Science and Technology Council (NSTC)
- 112-2314-B-075 -034 -MY3/National Science and Technology Council (NSTC)
- 112-2636-B-007-008/National Science and Technology Council (NSTC)
- 112-2314-B-002 -016/National Science and Technology Council (NSTC)
- 109-2314-B-075-044-MY3/National Science and Technology Council (NSTC)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

