Bacteriophages from human skin infecting coagulase-negative Staphylococcus: diversity, novelty and host resistance
- PMID: 38589670
- PMCID: PMC11001980
- DOI: 10.1038/s41598-024-59065-9
Bacteriophages from human skin infecting coagulase-negative Staphylococcus: diversity, novelty and host resistance
Abstract
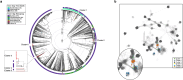
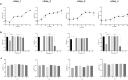
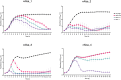
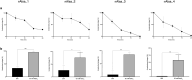
The human skin microbiome comprises diverse populations that differ temporally between body sites and individuals. The virome is a less studied component of the skin microbiome and the study of bacteriophages is required to increase knowledge of the modulation and stability of bacterial communities. Staphylococcus species are among the most abundant colonisers of skin and are associated with both health and disease yet the bacteriophages infecting the most abundant species on skin are less well studied. Here, we report the isolation and genome sequencing of 40 bacteriophages from human skin swabs that infect coagulase-negative Staphylococcus (CoNS) species, which extends our knowledge of phage diversity. Six genetic clusters of phages were identified with two clusters representing novel phages, one of which we characterise and name Alsa phage. We identified that Alsa phages have a greater ability to infect the species S. hominis that was otherwise infected less than other CoNS species by the isolated phages, indicating an undescribed barrier to phage infection that could be in part due to numerous restriction-modification systems. The extended diversity of Staphylococcus phages here enables further research to define their contribution to skin microbiome research and the mechanisms that limit phage infection.
Keywords: Bacteriophage; Comparative genomics; Defence mechanisms; Skin; Staphylococcus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Coagulase-Negative Staphylococci phages panorama: Genomic diversity and in vitro studies for a therapeutic use.Microbiol Res. 2025 Jan;290:127944. doi: 10.1016/j.micres.2024.127944. Epub 2024 Nov 9. Microbiol Res. 2025. PMID: 39550872 Review.
-
Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates.mBio. 2012 Sep 25;3(5):e00279-12. doi: 10.1128/mBio.00279-12. Print 2012. mBio. 2012. PMID: 23015740 Free PMC article.
-
Comparative Genomics of Three Novel Jumbo Bacteriophages Infecting Staphylococcus aureus.J Virol. 2021 Sep 9;95(19):e0239120. doi: 10.1128/JVI.02391-20. Epub 2021 Sep 9. J Virol. 2021. PMID: 34287047 Free PMC article.
-
Characterization of novel phages isolated in coagulase-negative staphylococci reveals evolutionary relationships with Staphylococcus aureus phages.J Bacteriol. 2012 Nov;194(21):5829-39. doi: 10.1128/JB.01085-12. Epub 2012 Aug 24. J Bacteriol. 2012. PMID: 22923589 Free PMC article.
-
The phages of staphylococci: critical catalysts in health and disease.Trends Microbiol. 2021 Dec;29(12):1117-1129. doi: 10.1016/j.tim.2021.04.008. Epub 2021 May 21. Trends Microbiol. 2021. PMID: 34030968 Free PMC article. Review.
Cited by
-
Bidirectional Mendelian Randomization Analysis to Study the Relationship Between Human Skin Microbiota and Radiation-Induced Skin Toxicity.Microorganisms. 2025 Jan 17;13(1):194. doi: 10.3390/microorganisms13010194. Microorganisms. 2025. PMID: 39858962 Free PMC article.
-
Recent Progress in Terrestrial Biota Derived Antibacterial Agents for Medical Applications.Molecules. 2024 Oct 15;29(20):4889. doi: 10.3390/molecules29204889. Molecules. 2024. PMID: 39459256 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

