Wnt, glucocorticoid and cellular prion protein cooperate to drive a mesenchymal phenotype with poor prognosis in colon cancer
- PMID: 38589873
- PMCID: PMC11003154
- DOI: 10.1186/s12967-024-05164-0
Wnt, glucocorticoid and cellular prion protein cooperate to drive a mesenchymal phenotype with poor prognosis in colon cancer
Abstract
Background: The mesenchymal subtype of colorectal cancer (CRC), associated with poor prognosis, is characterized by abundant expression of the cellular prion protein PrPC, which represents a candidate therapeutic target. How PrPC is induced in CRC remains elusive. This study aims to elucidate the signaling pathways governing PrPC expression and to shed light on the gene regulatory networks linked to PrPC.
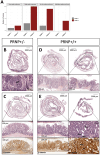
Methods: We performed in silico analyses on diverse datasets of in vitro, ex vivo and in vivo models of mouse CRC and patient cohorts. We mined ChIPseq studies and performed promoter analysis. CRC cell lines were manipulated through genetic and pharmacological approaches. We created mice combining conditional inactivation of Apc in intestinal epithelial cells and overexpression of the human prion protein gene PRNP. Bio-informatic analyses were carried out in two randomized control trials totalizing over 3000 CRC patients.
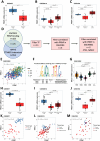
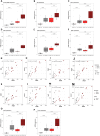
Results: In silico analyses combined with cell-based assays identified the Wnt-β-catenin and glucocorticoid pathways as upstream regulators of PRNP expression, with subtle differences between mouse and human. We uncover multiple feedback loops between PrPC and these two pathways, which translate into an aggravation of CRC pathogenesis in mouse. In stage III CRC patients, the signature defined by PRNP-CTNNB1-NR3C1, encoding PrPC, β-catenin and the glucocorticoid receptor respectively, is overrepresented in the poor-prognosis, mesenchymal subtype and associates with reduced time to recurrence.
Conclusions: An unleashed PrPC-dependent vicious circle is pathognomonic of poor prognosis, mesenchymal CRC. Patients from this aggressive subtype of CRC may benefit from therapies targeting the PRNP-CTNNB1-NR3C1 axis.
Keywords: Colon cancer; Glucocorticoid receptor; Molecular classification; Prion protein; Wnt-β-catenin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests in the present study.
Figures





References
-
- Parsons MJ, Tammela T, Dow LE. WNT as a driver and dependency in cancer. Cancer Discov. 2021;11:2413–2429. doi: 10.1158/2159-8290.CD-21-0190. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

