Mosaic derivative chromosomes at chorionic villi (CV) sampling are expression of genomic instability and precursors of cryptic disease-causing rearrangements: report of further four cases
- PMID: 38589928
- PMCID: PMC11003029
- DOI: 10.1186/s13039-024-00675-3
Mosaic derivative chromosomes at chorionic villi (CV) sampling are expression of genomic instability and precursors of cryptic disease-causing rearrangements: report of further four cases
Abstract
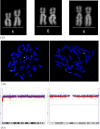
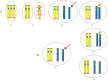
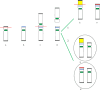
Mosaic chromosomal anomalies arising in the product of conception and the final fetal chromosomal arrangement are expression of complex biological mechanisms. The rescue of unbalanced chromosome with selection of the most viable cell line/s in the embryo and the unfavourable imbalances in placental tissues was documented in our previous paper and in the literature. We report four additional cases with mosaic derivative chromosomes in different feto-placental tissues, further showing the instability of an intermediate gross imbalance as a frequent mechanism of de novo cryptic deletions and duplications. In conclusion we underline how the extensive remodeling of unbalanced chromosomes in placental tissues represents the 'backstage' of de novo structural rearrangements, as the early phases of a long selection process that the genome undergo during embryogenesis.
Keywords: Chorionic villi; Cryptic rearrangement; Genomic instability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bonaglia MC, Kurtas NE, Errichiello E, Bertuzzo S, Beri S, Mehrjouy MM, Provenzano A, Vergani D, Pecile V, Novara F, Reho P, Di Giacomo MC, Discepoli G, Giorda R, Aldred MA, Santos-Rebouças CB, Goncalves AP, Abuelo DN, Giglio S, Ricca I, Franchi F, Patsalis P, Sismani C, Morí MA, Nevado J, Tommerup N, Zuffardi O. De novo unbalanced translocations have a complex history/aetiology. Hum Genet. 2018;137:817–829. doi: 10.1007/s00439-018-1941-9. - DOI - PubMed
LinkOut - more resources
Full Text Sources

